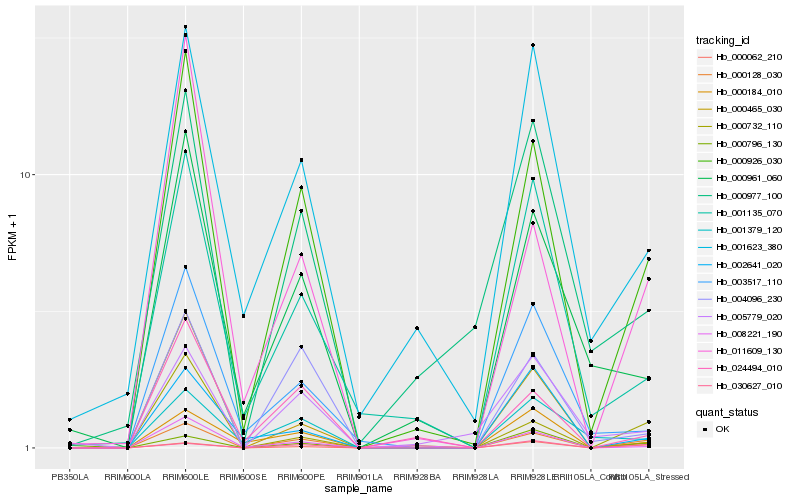
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000128_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000926_030 |
0.1151174365 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000062_210 |
0.175317367 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Pyrus x bretschneideri] |
| 4 |
Hb_030627_010 |
0.190139834 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 [Sesamum indicum] |
| 5 |
Hb_000732_110 |
0.2154154148 |
- |
- |
hypothetical protein JCGZ_21339 [Jatropha curcas] |
| 6 |
Hb_004096_230 |
0.2181797865 |
- |
- |
PREDICTED: uncharacterized protein LOC105638704 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000184_010 |
0.2224403024 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 8 |
Hb_011609_130 |
0.2369045572 |
- |
- |
- |
| 9 |
Hb_001135_070 |
0.2385135005 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 10 |
Hb_024494_010 |
0.2392786666 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 11 |
Hb_001623_380 |
0.2439529182 |
- |
- |
PREDICTED: 7-hydroxymethyl chlorophyll a reductase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000961_060 |
0.2485587658 |
- |
- |
uvb-resistance protein uvr8, putative [Ricinus communis] |
| 13 |
Hb_003517_110 |
0.2486124958 |
- |
- |
PREDICTED: uncharacterized protein LOC105633058 [Jatropha curcas] |
| 14 |
Hb_001379_120 |
0.2487210713 |
- |
- |
hypothetical protein POPTR_0001s29730g [Populus trichocarpa] |
| 15 |
Hb_000977_100 |
0.2500743133 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: acetolactate synthase small subunit 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000796_130 |
0.2524666165 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 17 |
Hb_008221_190 |
0.2525925944 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 18 |
Hb_002641_020 |
0.2539314931 |
- |
- |
3'-5' exonuclease, putative [Ricinus communis] |
| 19 |
Hb_005779_020 |
0.2559636428 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000465_030 |
0.2577600183 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |