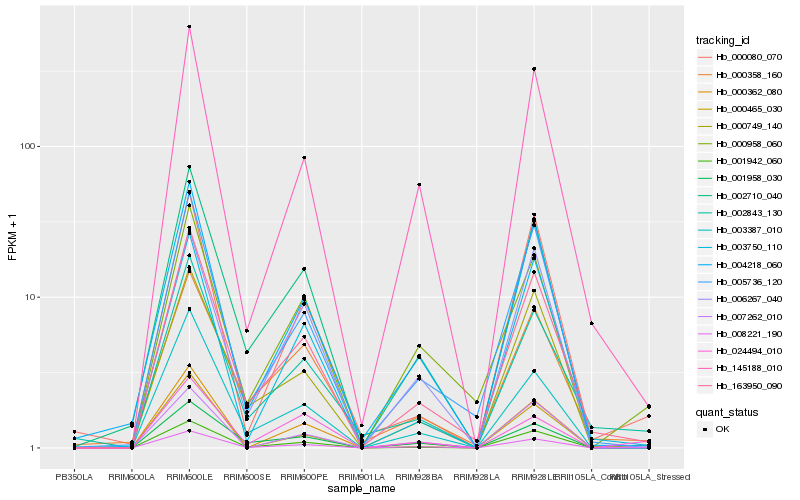
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008221_190 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 2 |
Hb_000958_060 |
0.105463029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_163950_090 |
0.1149126937 |
- |
- |
PREDICTED: traB domain-containing protein isoform X1 [Jatropha curcas] |
| 4 |
Hb_004218_060 |
0.1220854766 |
- |
- |
annexin [Manihot esculenta] |
| 5 |
Hb_145188_010 |
0.1227926443 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 6 |
Hb_002710_040 |
0.126917587 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 7 |
Hb_001942_060 |
0.1310543839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002843_130 |
0.132588217 |
- |
- |
PREDICTED: GRF1-interacting factor 2-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_006267_040 |
0.1366932511 |
- |
- |
PREDICTED: ABC transporter G family member 32 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000465_030 |
0.1366956893 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_005736_120 |
0.1376242615 |
- |
- |
PREDICTED: carboxyvinyl-carboxyphosphonate phosphorylmutase, chloroplastic [Gossypium raimondii] |
| 12 |
Hb_001958_030 |
0.138603354 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 13 |
Hb_000362_080 |
0.1405035931 |
- |
- |
hypothetical protein EUTSA_v10027329mg, partial [Eutrema salsugineum] |
| 14 |
Hb_003387_010 |
0.1434816859 |
- |
- |
PREDICTED: protein kinase PINOID 2 [Gossypium raimondii] |
| 15 |
Hb_024494_010 |
0.1453754603 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 16 |
Hb_000358_160 |
0.1457572928 |
- |
- |
PREDICTED: uncharacterized protein LOC105637769 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000749_140 |
0.1464199083 |
- |
- |
PREDICTED: 2-succinylbenzoate--CoA ligase, chloroplastic/peroxisomal [Jatropha curcas] |
| 18 |
Hb_007262_010 |
0.1469939802 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000080_070 |
0.1471681061 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_003750_110 |
0.1489258007 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |