| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
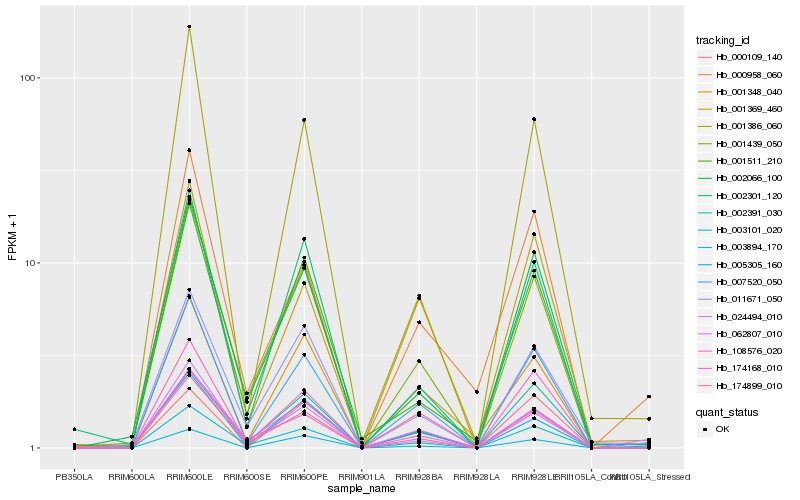
Hb_007520_050 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 2 |
Hb_174168_010 |
0.0856070774 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 3 |
Hb_003101_020 |
0.0981275091 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 4 |
Hb_001439_050 |
0.0984612081 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_062807_010 |
0.1053298925 |
- |
- |
hypothetical protein JCGZ_15896 [Jatropha curcas] |
| 6 |
Hb_001369_460 |
0.1074339762 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 7 |
Hb_005305_160 |
0.1173351281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002066_100 |
0.1220260393 |
- |
- |
PREDICTED: protein disulfide-isomerase SCO2 [Jatropha curcas] |
| 9 |
Hb_002391_030 |
0.1229878422 |
- |
- |
hypothetical protein CISIN_1g020187mg [Citrus sinensis] |
| 10 |
Hb_108576_020 |
0.1251828635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_174899_010 |
0.1262842428 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 12 |
Hb_000958_060 |
0.1274980377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003894_170 |
0.1279129742 |
transcription factor |
TF Family: LOB |
hypothetical protein CICLE_v10021992mg [Citrus clementina] |
| 14 |
Hb_011671_050 |
0.1286649406 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 15 |
Hb_001348_040 |
0.1301464551 |
- |
- |
hypothetical protein VITISV_006651 [Vitis vinifera] |
| 16 |
Hb_024494_010 |
0.1312378996 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 17 |
Hb_000109_140 |
0.131481813 |
- |
- |
hypothetical protein CICLE_v100314482mg, partial [Citrus clementina] |
| 18 |
Hb_001386_060 |
0.1330875376 |
- |
- |
PREDICTED: acetolactate synthase 1, chloroplastic-like [Vitis vinifera] |
| 19 |
Hb_002301_120 |
0.1332951954 |
- |
- |
PREDICTED: uncharacterized protein LOC105649581 [Jatropha curcas] |
| 20 |
Hb_001511_210 |
0.1345507145 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |