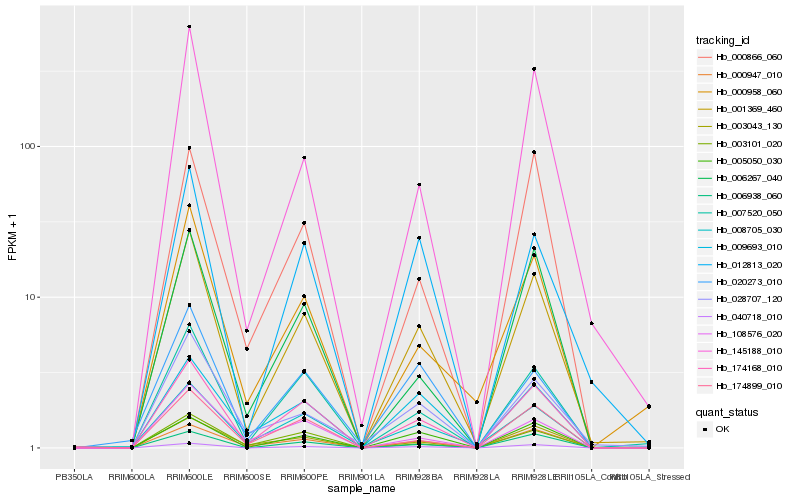
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_460 |
0.0 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 2 |
Hb_174168_010 |
0.077471542 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 3 |
Hb_040718_010 |
0.0968461303 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 4 |
Hb_007520_050 |
0.1074339762 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 5 |
Hb_000947_010 |
0.1154077195 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 6 |
Hb_006938_060 |
0.1216648945 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 7 |
Hb_003043_130 |
0.1230200573 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF2-like [Jatropha curcas] |
| 8 |
Hb_145188_010 |
0.1263694958 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 9 |
Hb_000958_060 |
0.1279326796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005050_030 |
0.127937746 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like [Jatropha curcas] |
| 11 |
Hb_012813_020 |
0.1282979164 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase, basic isoform-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_008705_030 |
0.1300336262 |
- |
- |
hypothetical protein JCGZ_13017 [Jatropha curcas] |
| 13 |
Hb_028707_120 |
0.1308459709 |
- |
- |
PREDICTED: uncharacterized protein LOC105645799 isoform X2 [Jatropha curcas] |
| 14 |
Hb_009693_010 |
0.1438060346 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Gossypium raimondii] |
| 15 |
Hb_003101_020 |
0.146923634 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 16 |
Hb_006267_040 |
0.1469769896 |
- |
- |
PREDICTED: ABC transporter G family member 32 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000866_060 |
0.1542123258 |
- |
- |
putative glycerol-3-phosphate acyltransferase [Vernicia fordii] |
| 18 |
Hb_020273_010 |
0.1548441281 |
- |
- |
PREDICTED: beta-galactosidase 3 [Jatropha curcas] |
| 19 |
Hb_174899_010 |
0.1554226032 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 20 |
Hb_108576_020 |
0.1570304587 |
- |
- |
conserved hypothetical protein [Ricinus communis] |