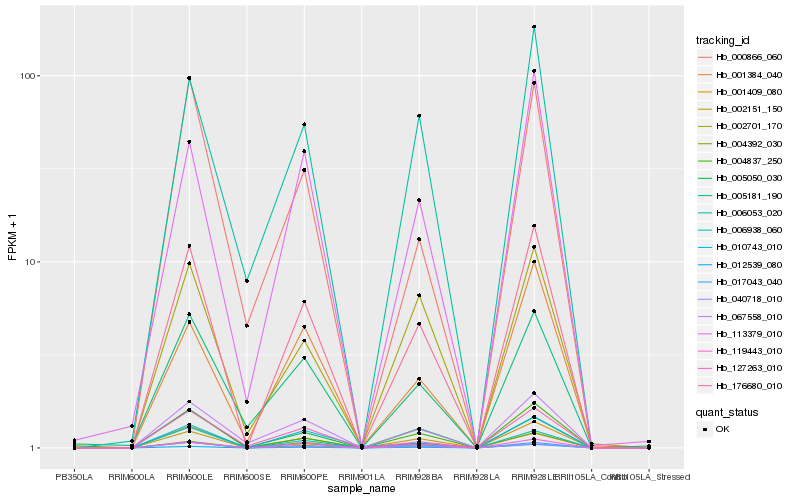
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_176680_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_22787 [Jatropha curcas] |
| 2 |
Hb_001409_080 |
0.0799190043 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Populus euphratica] |
| 3 |
Hb_006938_060 |
0.0934063083 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 4 |
Hb_119443_010 |
0.0949417054 |
- |
- |
hypothetical protein JCGZ_17348 [Jatropha curcas] |
| 5 |
Hb_067558_010 |
0.1148284527 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 6 |
Hb_012539_080 |
0.1184323053 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 7 |
Hb_040718_010 |
0.1191890492 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 8 |
Hb_127263_010 |
0.1285155489 |
- |
- |
hypothetical protein JCGZ_02828 [Jatropha curcas] |
| 9 |
Hb_113379_010 |
0.1285717817 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1 [Jatropha curcas] |
| 10 |
Hb_006053_020 |
0.1294094378 |
- |
- |
CAP [Theobroma cacao] |
| 11 |
Hb_002151_150 |
0.1313494691 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |
| 12 |
Hb_004837_250 |
0.1315843724 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005050_030 |
0.1349521154 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like [Jatropha curcas] |
| 14 |
Hb_017043_040 |
0.1355727722 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 15 |
Hb_001384_040 |
0.136566939 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 16 |
Hb_005181_190 |
0.1367623832 |
- |
- |
PREDICTED: uncharacterized protein LOC105643550 [Jatropha curcas] |
| 17 |
Hb_002701_170 |
0.1377320332 |
- |
- |
PREDICTED: cytochrome P450 85A [Jatropha curcas] |
| 18 |
Hb_004392_030 |
0.138187625 |
- |
- |
oxidoreductase family protein [Populus trichocarpa] |
| 19 |
Hb_010743_010 |
0.1385009915 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_000866_060 |
0.1390367967 |
- |
- |
putative glycerol-3-phosphate acyltransferase [Vernicia fordii] |