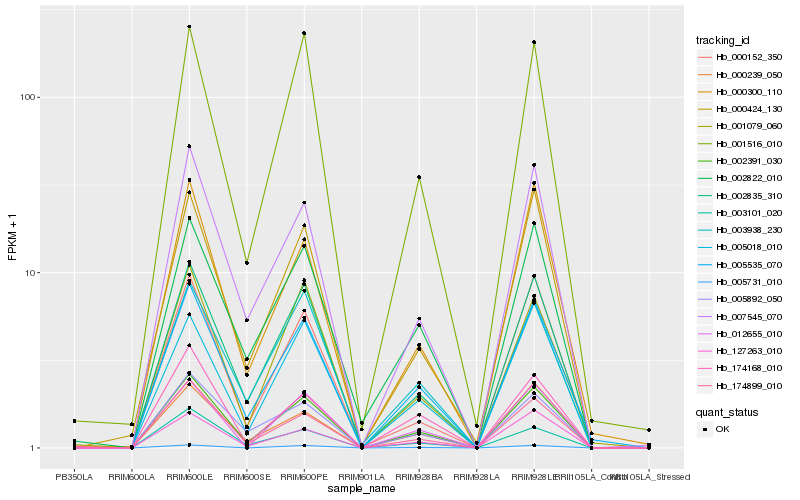
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002391_030 |
0.0 |
- |
- |
hypothetical protein CISIN_1g020187mg [Citrus sinensis] |
| 2 |
Hb_005535_070 |
0.0599995643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002835_310 |
0.0638405086 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_007545_070 |
0.078571539 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 306 [Populus euphratica] |
| 5 |
Hb_000424_130 |
0.0886467999 |
- |
- |
PREDICTED: monoacylglycerol lipase abhd6-B-like [Jatropha curcas] |
| 6 |
Hb_001516_010 |
0.0888354416 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 7 |
Hb_174899_010 |
0.0914102962 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 8 |
Hb_000239_050 |
0.0914775567 |
- |
- |
PREDICTED: cyclin-D5-1-like [Jatropha curcas] |
| 9 |
Hb_005018_010 |
0.0917057985 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 10 |
Hb_127263_010 |
0.0941887239 |
- |
- |
hypothetical protein JCGZ_02828 [Jatropha curcas] |
| 11 |
Hb_174168_010 |
0.0979545606 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 12 |
Hb_001079_060 |
0.0982066427 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003938_230 |
0.0988821361 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 14 |
Hb_003101_020 |
0.0994642394 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 15 |
Hb_002822_010 |
0.1000079484 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Populus euphratica] |
| 16 |
Hb_012655_010 |
0.1009308685 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 17 |
Hb_000300_110 |
0.102566371 |
- |
- |
hypothetical protein JCGZ_02651 [Jatropha curcas] |
| 18 |
Hb_005731_010 |
0.1035157781 |
- |
- |
hypothetical protein VITISV_030941 [Vitis vinifera] |
| 19 |
Hb_005892_050 |
0.1040776515 |
- |
- |
PREDICTED: uncharacterized protein LOC105632175 [Jatropha curcas] |
| 20 |
Hb_000152_350 |
0.1057768068 |
- |
- |
PREDICTED: enolase [Jatropha curcas] |