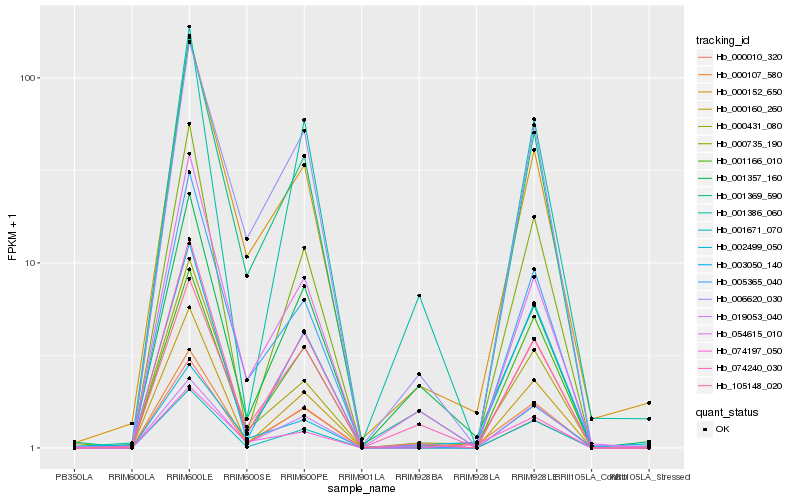
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_580 |
0.0 |
- |
- |
PREDICTED: delta(24)-sterol reductase [Jatropha curcas] |
| 2 |
Hb_003050_140 |
0.1019064625 |
- |
- |
PREDICTED: potassium channel KAT1 [Jatropha curcas] |
| 3 |
Hb_000152_650 |
0.1064408619 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 4 |
Hb_001369_590 |
0.1093871653 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 5 |
Hb_105148_020 |
0.112752249 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 6 |
Hb_001357_160 |
0.1148645537 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_074240_030 |
0.1164032043 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 3-like [Jatropha curcas] |
| 8 |
Hb_006620_030 |
0.121453873 |
- |
- |
nitrate reductase, putative [Ricinus communis] |
| 9 |
Hb_000010_320 |
0.1234825849 |
- |
- |
PREDICTED: protein STICHEL-like 2 [Jatropha curcas] |
| 10 |
Hb_001671_070 |
0.1244677733 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_000160_260 |
0.1249493219 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 12 |
Hb_001386_060 |
0.1256138856 |
- |
- |
PREDICTED: acetolactate synthase 1, chloroplastic-like [Vitis vinifera] |
| 13 |
Hb_000735_190 |
0.1263580039 |
- |
- |
hypothetical protein POPTR_0006s19340g [Populus trichocarpa] |
| 14 |
Hb_001166_010 |
0.1277789281 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 15 |
Hb_054615_010 |
0.1290840773 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 16 |
Hb_000431_080 |
0.1300214449 |
- |
- |
PREDICTED: uncharacterized protein LOC105632547 [Jatropha curcas] |
| 17 |
Hb_019053_040 |
0.1307260846 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_005365_040 |
0.131018664 |
- |
- |
hypothetical protein JCGZ_15100 [Jatropha curcas] |
| 19 |
Hb_074197_050 |
0.1314538516 |
- |
- |
PREDICTED: uncharacterized protein LOC105645952 [Jatropha curcas] |
| 20 |
Hb_002499_050 |
0.1315408516 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |