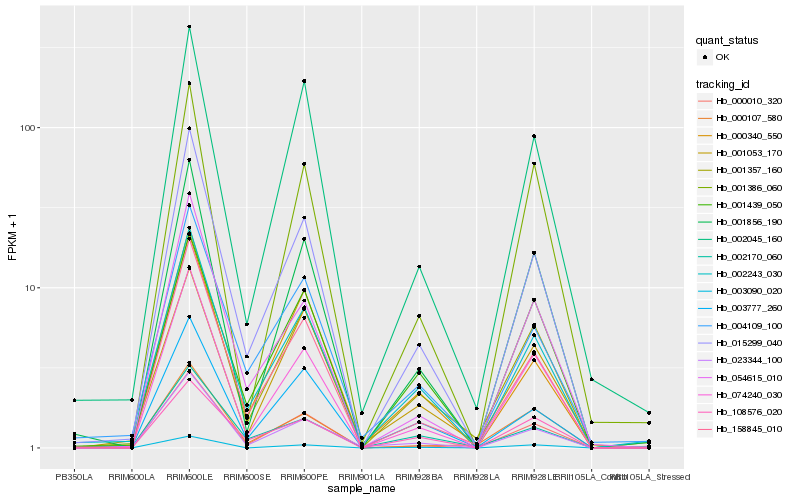
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001357_160 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002243_030 |
0.0754040486 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 3-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_074240_030 |
0.0772905665 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 3-like [Jatropha curcas] |
| 4 |
Hb_015299_040 |
0.0789074048 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 [Jatropha curcas] |
| 5 |
Hb_158845_010 |
0.0873216866 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor At1g16060 [Jatropha curcas] |
| 6 |
Hb_001856_190 |
0.0890060122 |
- |
- |
SP1L, putative [Ricinus communis] |
| 7 |
Hb_000010_320 |
0.0955264014 |
- |
- |
PREDICTED: protein STICHEL-like 2 [Jatropha curcas] |
| 8 |
Hb_023344_100 |
0.0985543441 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.2 [Jatropha curcas] |
| 9 |
Hb_003777_260 |
0.103029345 |
- |
- |
PREDICTED: uncharacterized protein LOC105640927 isoform X4 [Jatropha curcas] |
| 10 |
Hb_003090_020 |
0.1047045296 |
- |
- |
PREDICTED: uncharacterized protein LOC105646662 [Jatropha curcas] |
| 11 |
Hb_001386_060 |
0.1068672397 |
- |
- |
PREDICTED: acetolactate synthase 1, chloroplastic-like [Vitis vinifera] |
| 12 |
Hb_002045_160 |
0.111602775 |
- |
- |
PREDICTED: DJ-1 protein homolog E-like [Jatropha curcas] |
| 13 |
Hb_054615_010 |
0.1140833716 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 14 |
Hb_000107_580 |
0.1148645537 |
- |
- |
PREDICTED: delta(24)-sterol reductase [Jatropha curcas] |
| 15 |
Hb_001439_050 |
0.1162227331 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 16 |
Hb_004109_100 |
0.1183101711 |
- |
- |
Interactor of constitutive active rops 1 isoform 1 [Theobroma cacao] |
| 17 |
Hb_002170_060 |
0.1190969621 |
- |
- |
PREDICTED: mitogen-activated protein kinase-binding protein 1 [Jatropha curcas] |
| 18 |
Hb_001053_170 |
0.1230524131 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 19 |
Hb_108576_020 |
0.1232610764 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000340_550 |
0.1256088963 |
- |
- |
PREDICTED: rho GTPase-activating protein 2 [Jatropha curcas] |