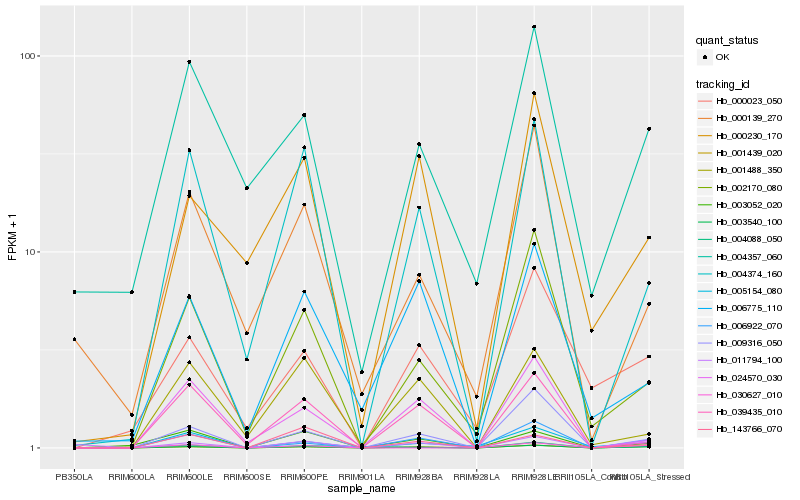
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003540_100 |
0.0 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 2 |
Hb_001439_020 |
0.0606511797 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 3 |
Hb_006922_070 |
0.1647609932 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 4 |
Hb_009316_050 |
0.1954339677 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 5 |
Hb_004374_160 |
0.210831444 |
transcription factor |
TF Family: bHLH |
hypothetical protein JCGZ_20587 [Jatropha curcas] |
| 6 |
Hb_002170_080 |
0.2196184114 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 7 |
Hb_011794_100 |
0.2221390983 |
- |
- |
PREDICTED: seven transmembrane domain-containing tyrosine-protein kinase 1-like [Jatropha curcas] |
| 8 |
Hb_005154_080 |
0.2283863744 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640764 [Jatropha curcas] |
| 9 |
Hb_030627_010 |
0.2349689738 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 [Sesamum indicum] |
| 10 |
Hb_003052_020 |
0.236358296 |
- |
- |
PREDICTED: chaperonin 60 subunit beta 4, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_000023_050 |
0.2478272617 |
- |
- |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 12 |
Hb_143766_070 |
0.2553274196 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 13 |
Hb_004357_060 |
0.2573673941 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 14 |
Hb_000230_170 |
0.2588685605 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1 [Jatropha curcas] |
| 15 |
Hb_006775_110 |
0.2637873539 |
- |
- |
PREDICTED: uncharacterized protein LOC105649734 [Jatropha curcas] |
| 16 |
Hb_039435_010 |
0.2669288643 |
- |
- |
hypothetical protein SORBIDRAFT_10g025530 [Sorghum bicolor] |
| 17 |
Hb_004088_050 |
0.2681832119 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 18 |
Hb_024570_030 |
0.2683311813 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_000139_270 |
0.27011758 |
- |
- |
hypothetical protein POPTR_0008s14890g [Populus trichocarpa] |
| 20 |
Hb_001488_350 |
0.2748161036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |