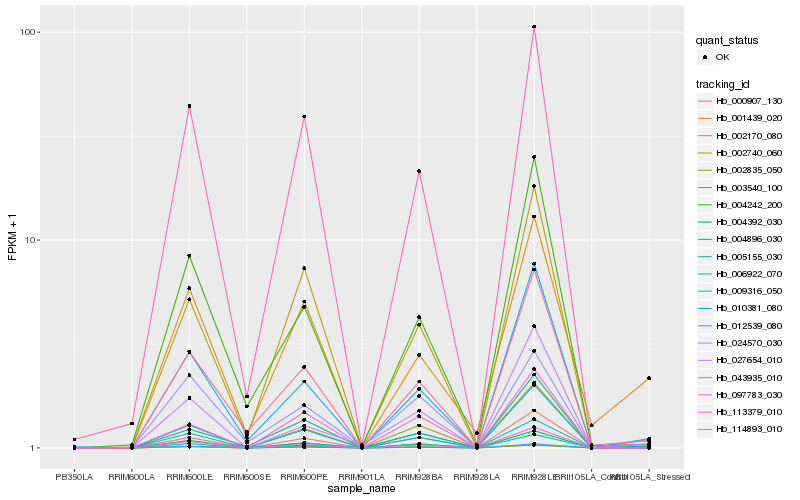
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009316_050 |
0.0 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 2 |
Hb_002170_080 |
0.1425033825 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 3 |
Hb_006922_070 |
0.1513953212 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 4 |
Hb_027654_010 |
0.1641313167 |
- |
- |
Isoamyl acetate-hydrolyzing esterase, putative [Ricinus communis] |
| 5 |
Hb_010381_080 |
0.177896961 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_000907_130 |
0.1825493499 |
- |
- |
- |
| 7 |
Hb_002835_050 |
0.1838648069 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 8 |
Hb_012539_080 |
0.1849842608 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 9 |
Hb_043935_010 |
0.1863181503 |
- |
- |
Receptor protein kinase-like protein [Medicago truncatula] |
| 10 |
Hb_002740_060 |
0.1911836751 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 11 |
Hb_005155_030 |
0.1930209194 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 12 |
Hb_004392_030 |
0.1947710036 |
- |
- |
oxidoreductase family protein [Populus trichocarpa] |
| 13 |
Hb_003540_100 |
0.1954339677 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 14 |
Hb_001439_020 |
0.19594997 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 15 |
Hb_113379_010 |
0.1970039612 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1 [Jatropha curcas] |
| 16 |
Hb_024570_030 |
0.1971231641 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_004242_200 |
0.1983019798 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 18 |
Hb_114893_010 |
0.1983802148 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 19 |
Hb_004896_030 |
0.2050207292 |
- |
- |
hypothetical protein VITISV_007475 [Vitis vinifera] |
| 20 |
Hb_097783_030 |
0.2064766624 |
- |
- |
PREDICTED: probable peroxidase 61 [Jatropha curcas] |