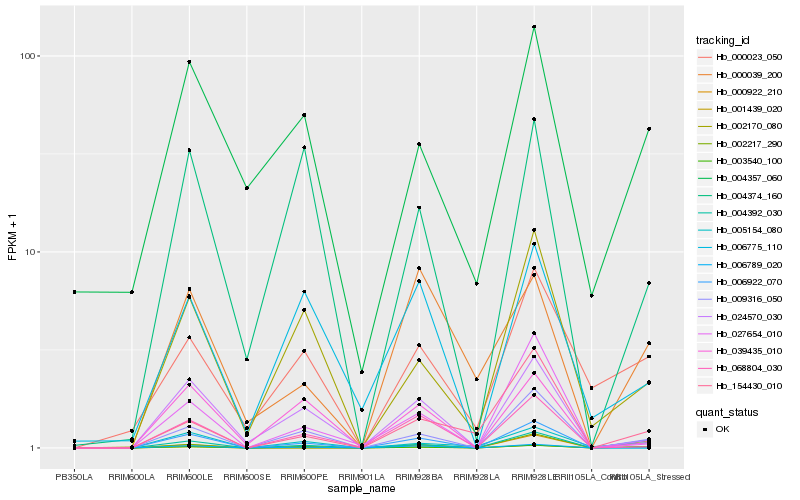
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006922_070 |
0.0 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 2 |
Hb_001439_020 |
0.1434208285 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 3 |
Hb_009316_050 |
0.1513953212 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 4 |
Hb_003540_100 |
0.1647609932 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 5 |
Hb_024570_030 |
0.2119058907 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_005154_080 |
0.2134248342 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640764 [Jatropha curcas] |
| 7 |
Hb_002170_080 |
0.2202592119 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 8 |
Hb_068804_030 |
0.2336321783 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 9 |
Hb_027654_010 |
0.2398466153 |
- |
- |
Isoamyl acetate-hydrolyzing esterase, putative [Ricinus communis] |
| 10 |
Hb_000023_050 |
0.2423050264 |
- |
- |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 11 |
Hb_154430_010 |
0.2473804261 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X2 [Jatropha curcas] |
| 12 |
Hb_004374_160 |
0.2530253863 |
transcription factor |
TF Family: bHLH |
hypothetical protein JCGZ_20587 [Jatropha curcas] |
| 13 |
Hb_039435_010 |
0.2540326596 |
- |
- |
hypothetical protein SORBIDRAFT_10g025530 [Sorghum bicolor] |
| 14 |
Hb_006775_110 |
0.258677048 |
- |
- |
PREDICTED: uncharacterized protein LOC105649734 [Jatropha curcas] |
| 15 |
Hb_004392_030 |
0.2599102069 |
- |
- |
oxidoreductase family protein [Populus trichocarpa] |
| 16 |
Hb_002217_290 |
0.2630965967 |
- |
- |
hypothetical protein JCGZ_19038 [Jatropha curcas] |
| 17 |
Hb_000922_210 |
0.2633906968 |
- |
- |
PREDICTED: 50S ribosomal protein L12, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000039_200 |
0.2636818868 |
- |
- |
hypothetical protein JCGZ_21346 [Jatropha curcas] |
| 19 |
Hb_004357_060 |
0.2637672974 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 20 |
Hb_006789_020 |
0.2638995461 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: wall-associated receptor kinase-like 22 [Camelina sativa] |