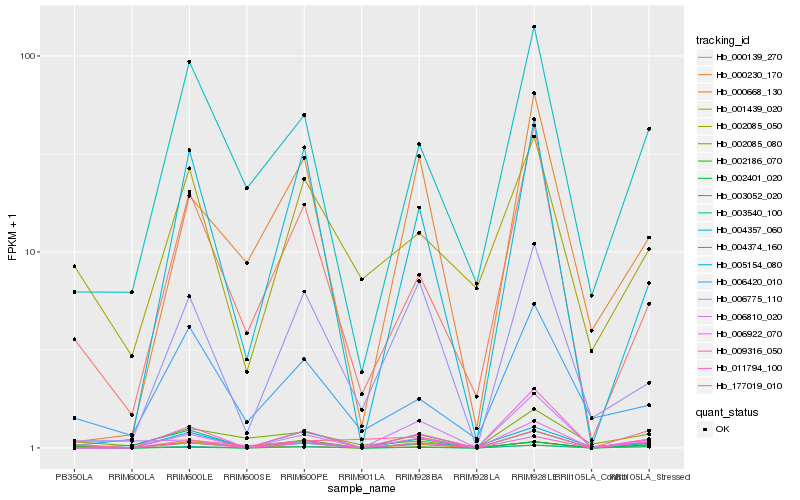
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011794_100 |
0.0 |
- |
- |
PREDICTED: seven transmembrane domain-containing tyrosine-protein kinase 1-like [Jatropha curcas] |
| 2 |
Hb_001439_020 |
0.2185898447 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 3 |
Hb_003540_100 |
0.2221390983 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 4 |
Hb_002401_020 |
0.24502227 |
- |
- |
PREDICTED: protein argonaute 16 isoform X2 [Jatropha curcas] |
| 5 |
Hb_006922_070 |
0.2798014943 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 6 |
Hb_000139_270 |
0.2873176071 |
- |
- |
hypothetical protein POPTR_0008s14890g [Populus trichocarpa] |
| 7 |
Hb_000668_130 |
0.2950891203 |
- |
- |
- |
| 8 |
Hb_002186_070 |
0.2959373984 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_002085_050 |
0.2960720511 |
- |
- |
PREDICTED: beta-galactosidase 8 [Jatropha curcas] |
| 10 |
Hb_004357_060 |
0.3000456213 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 11 |
Hb_004374_160 |
0.3003310486 |
transcription factor |
TF Family: bHLH |
hypothetical protein JCGZ_20587 [Jatropha curcas] |
| 12 |
Hb_005154_080 |
0.3011028165 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640764 [Jatropha curcas] |
| 13 |
Hb_009316_050 |
0.3033745194 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 14 |
Hb_006775_110 |
0.3078746174 |
- |
- |
PREDICTED: uncharacterized protein LOC105649734 [Jatropha curcas] |
| 15 |
Hb_002085_080 |
0.3080627105 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 68-like [Jatropha curcas] |
| 16 |
Hb_006810_020 |
0.3141948076 |
- |
- |
PREDICTED: NADPH:quinone oxidoreductase-like [Jatropha curcas] |
| 17 |
Hb_000230_170 |
0.3142024224 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1 [Jatropha curcas] |
| 18 |
Hb_003052_020 |
0.3148350691 |
- |
- |
PREDICTED: chaperonin 60 subunit beta 4, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_177019_010 |
0.3168216838 |
- |
- |
Uncharacterized protein TCM_004012 [Theobroma cacao] |
| 20 |
Hb_006420_010 |
0.3206064257 |
- |
- |
PREDICTED: uncharacterized protein LOC105649815 [Jatropha curcas] |