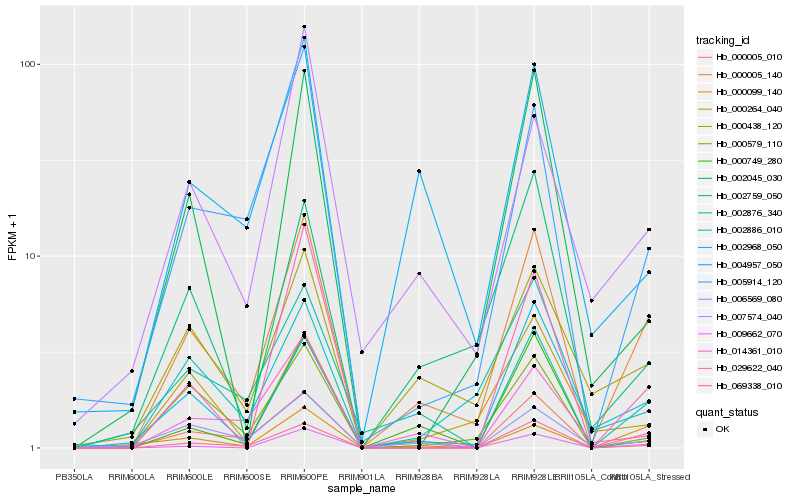
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000005_140 |
0.0 |
transcription factor |
TF Family: C2H2 |
unknown [Populus trichocarpa] |
| 2 |
Hb_069338_010 |
0.1735542622 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000438_120 |
0.1812209122 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 4 |
Hb_009662_070 |
0.1859958872 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 5 |
Hb_005914_120 |
0.1948146115 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 6 |
Hb_002968_050 |
0.1958506094 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 7 |
Hb_002759_050 |
0.202868629 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |
| 8 |
Hb_014361_010 |
0.2042195949 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 9 |
Hb_000579_110 |
0.2052674366 |
- |
- |
PREDICTED: transmembrane protein 136 [Jatropha curcas] |
| 10 |
Hb_006569_080 |
0.2066621708 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_024559 [Vitis vinifera] |
| 11 |
Hb_000099_140 |
0.2071140581 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 12 |
Hb_004957_050 |
0.2115489016 |
- |
- |
hypothetical protein POPTR_0010s12360g [Populus trichocarpa] |
| 13 |
Hb_002876_340 |
0.2118677715 |
- |
- |
hypothetical protein POPTR_0013s11770g [Populus trichocarpa] |
| 14 |
Hb_002886_010 |
0.2151418765 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000005_010 |
0.2152277646 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 16 |
Hb_029622_040 |
0.2152775546 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007574_040 |
0.2154407978 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 18 |
Hb_000264_040 |
0.2213921799 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 19 |
Hb_000749_280 |
0.2241215512 |
- |
- |
PREDICTED: uncharacterized protein LOC105138169 [Populus euphratica] |
| 20 |
Hb_002045_030 |
0.2246703576 |
transcription factor |
TF Family: AUX/IAA |
hypothetical protein CISIN_1g047687mg [Citrus sinensis] |