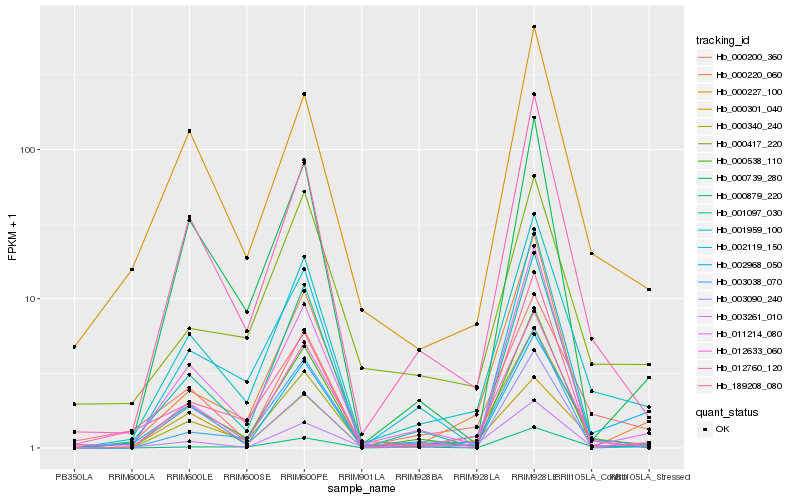
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001959_100 |
0.0 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 2 |
Hb_000340_240 |
0.0823941945 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_012760_120 |
0.1094529783 |
- |
- |
PREDICTED: peroxidase 64-like [Jatropha curcas] |
| 4 |
Hb_000200_360 |
0.111515519 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 5 |
Hb_000538_110 |
0.1180141216 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 6 |
Hb_000227_100 |
0.1274597416 |
- |
- |
PREDICTED: uncharacterized protein LOC105631933 [Jatropha curcas] |
| 7 |
Hb_011214_080 |
0.12884639 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 8 |
Hb_012633_060 |
0.1361107738 |
- |
- |
laccase, putative [Ricinus communis] |
| 9 |
Hb_003261_010 |
0.1401065789 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 10 |
Hb_000739_280 |
0.1445043204 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_000220_060 |
0.1470366544 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 15 [Jatropha curcas] |
| 12 |
Hb_001097_030 |
0.1484893242 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 13 |
Hb_000301_040 |
0.1506439727 |
- |
- |
PREDICTED: uncharacterized protein LOC105639235 [Jatropha curcas] |
| 14 |
Hb_002119_150 |
0.1526961846 |
- |
- |
hypothetical protein POPTR_0008s13870g [Populus trichocarpa] |
| 15 |
Hb_003090_240 |
0.1561199536 |
- |
- |
hypothetical protein JCGZ_25095 [Jatropha curcas] |
| 16 |
Hb_189208_080 |
0.1568643104 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 11-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_003038_070 |
0.1573862045 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 18 |
Hb_000879_220 |
0.1577697006 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 19 |
Hb_002968_050 |
0.1585217839 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 20 |
Hb_000417_220 |
0.1590665428 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Cucumis melo] |