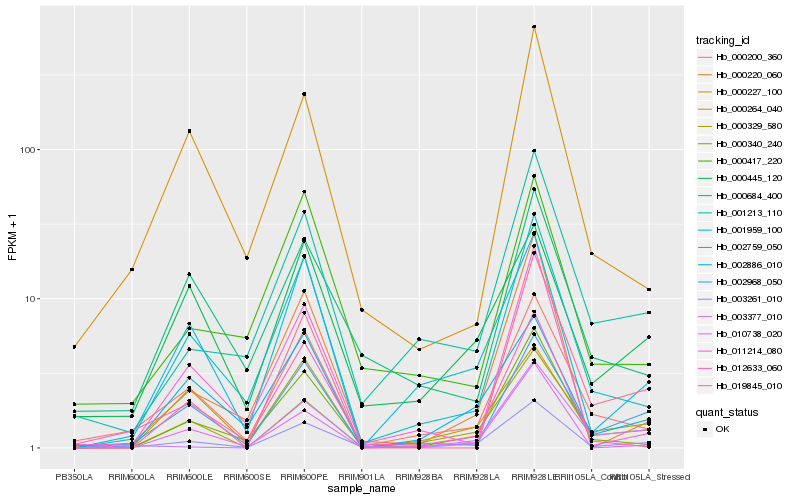
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003261_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 2 |
Hb_001959_100 |
0.1401065789 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 3 |
Hb_000340_240 |
0.1415935772 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000200_360 |
0.1523838008 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 5 |
Hb_000220_060 |
0.1550557137 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 15 [Jatropha curcas] |
| 6 |
Hb_000329_580 |
0.1569346344 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_002968_050 |
0.1576342754 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 8 |
Hb_001213_110 |
0.1603273998 |
- |
- |
SOUL heme-binding family protein [Populus trichocarpa] |
| 9 |
Hb_002759_050 |
0.163361858 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |
| 10 |
Hb_010738_020 |
0.1757595312 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 11 |
Hb_012633_060 |
0.1826224103 |
- |
- |
laccase, putative [Ricinus communis] |
| 12 |
Hb_003377_010 |
0.1860632473 |
- |
- |
hypothetical protein POPTR_0016s06140g, partial [Populus trichocarpa] |
| 13 |
Hb_000227_100 |
0.1864909848 |
- |
- |
PREDICTED: uncharacterized protein LOC105631933 [Jatropha curcas] |
| 14 |
Hb_002886_010 |
0.1867275014 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_019845_010 |
0.1889075745 |
- |
- |
protease inhibitor protein [Hevea brasiliensis] |
| 16 |
Hb_000417_220 |
0.1938830586 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Cucumis melo] |
| 17 |
Hb_011214_080 |
0.194409771 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 18 |
Hb_000264_040 |
0.1944776074 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 19 |
Hb_000445_120 |
0.1950268511 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_000684_400 |
0.2006482226 |
- |
- |
PREDICTED: uncharacterized protein LOC105642159 [Jatropha curcas] |