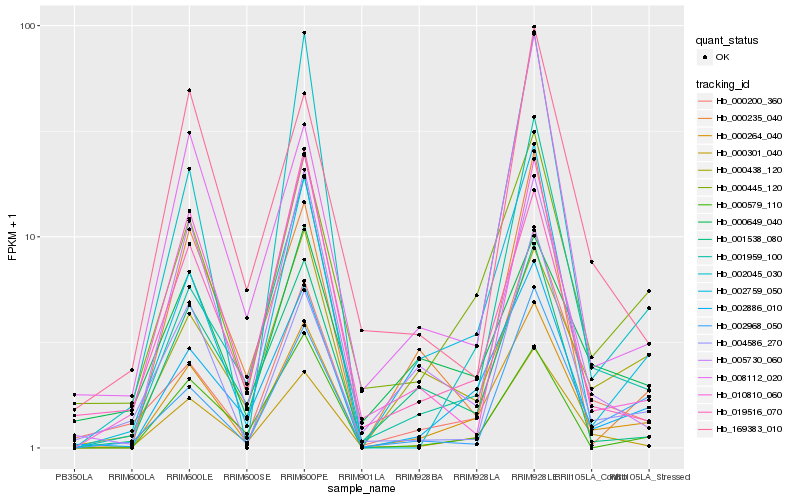
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000264_040 |
0.0 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 2 |
Hb_002886_010 |
0.089123531 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000445_120 |
0.1114219274 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_002759_050 |
0.1205535 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |
| 5 |
Hb_000301_040 |
0.1241476159 |
- |
- |
PREDICTED: uncharacterized protein LOC105639235 [Jatropha curcas] |
| 6 |
Hb_005730_060 |
0.1609323162 |
- |
- |
PREDICTED: inorganic pyrophosphatase 1-like [Jatropha curcas] |
| 7 |
Hb_000200_360 |
0.1647508427 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 8 |
Hb_002968_050 |
0.1706391941 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 9 |
Hb_000438_120 |
0.1728964328 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 10 |
Hb_001538_080 |
0.1751469771 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000579_110 |
0.1772891637 |
- |
- |
PREDICTED: transmembrane protein 136 [Jatropha curcas] |
| 12 |
Hb_001959_100 |
0.1777649953 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 13 |
Hb_010810_060 |
0.1791111085 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 14 |
Hb_004586_270 |
0.1794600607 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_008112_020 |
0.1849811592 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000235_040 |
0.1866414668 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 isoform X1 [Populus euphratica] |
| 17 |
Hb_000649_040 |
0.1875675478 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |
| 18 |
Hb_002045_030 |
0.1909561819 |
transcription factor |
TF Family: AUX/IAA |
hypothetical protein CISIN_1g047687mg [Citrus sinensis] |
| 19 |
Hb_019516_070 |
0.192602042 |
- |
- |
PREDICTED: uncharacterized protein LOC105634258 [Jatropha curcas] |
| 20 |
Hb_169383_010 |
0.1933084143 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |