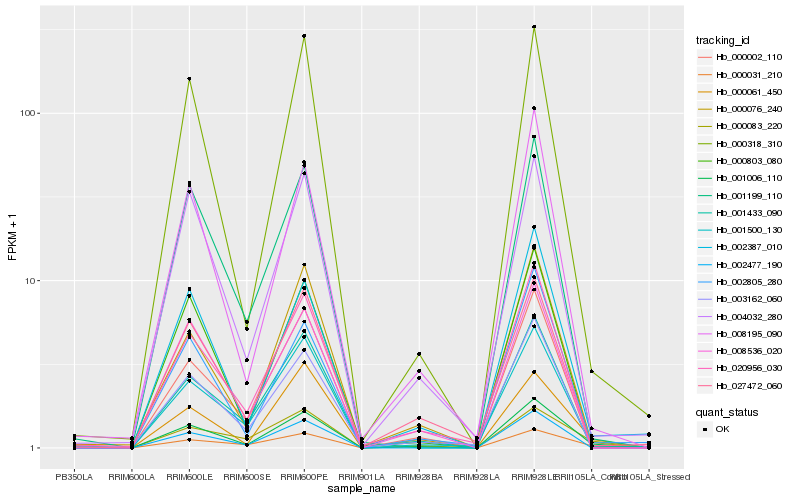
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001006_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634212 [Jatropha curcas] |
| 2 |
Hb_000031_210 |
0.1169345287 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_001500_130 |
0.1263347472 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 4 |
Hb_000083_220 |
0.1275957976 |
- |
- |
- |
| 5 |
Hb_002387_010 |
0.1280467049 |
- |
- |
PREDICTED: protein STAY-GREEN LIKE, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000076_240 |
0.1291148781 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 7 |
Hb_000318_310 |
0.1295258666 |
- |
- |
hypothetical protein POPTR_0009s11190g [Populus trichocarpa] |
| 8 |
Hb_000002_110 |
0.1323349271 |
- |
- |
hypothetical protein EUGRSUZ_B02140 [Eucalyptus grandis] |
| 9 |
Hb_020956_030 |
0.1323585887 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 10 |
Hb_008195_090 |
0.1329488412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000803_080 |
0.1330218111 |
desease resistance |
Gene Name: ABC_tran |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 12 |
Hb_004032_280 |
0.1330244427 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 13 |
Hb_001433_090 |
0.1331096697 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 14 |
Hb_008536_020 |
0.133158038 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 15 |
Hb_002805_280 |
0.134709886 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 16 |
Hb_000061_450 |
0.1348099179 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002477_190 |
0.1348641855 |
- |
- |
hypothetical protein EUGRSUZ_C03380, partial [Eucalyptus grandis] |
| 18 |
Hb_027472_060 |
0.1354324332 |
transcription factor |
TF Family: GRAS |
DELLA protein GAI1, putative [Ricinus communis] |
| 19 |
Hb_001199_110 |
0.1357064784 |
- |
- |
PREDICTED: uncharacterized protein ycf36 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003162_060 |
0.1363332044 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |