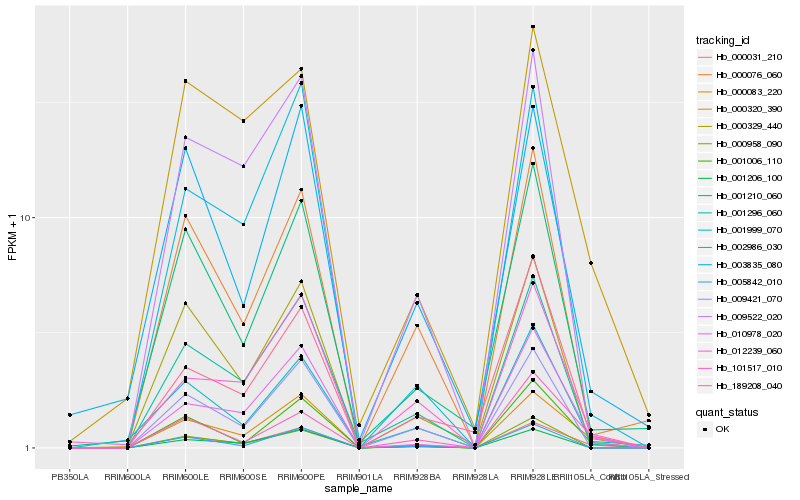
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000031_210 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_000083_220 |
0.0478048414 |
- |
- |
- |
| 3 |
Hb_010978_020 |
0.1130585043 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL12 [Jatropha curcas] |
| 4 |
Hb_001006_110 |
0.1169345287 |
- |
- |
PREDICTED: uncharacterized protein LOC105634212 [Jatropha curcas] |
| 5 |
Hb_001296_060 |
0.13105572 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 6 |
Hb_001999_070 |
0.1435182844 |
- |
- |
PREDICTED: ammonium transporter 1 member 3 [Jatropha curcas] |
| 7 |
Hb_002986_030 |
0.147745898 |
- |
- |
hypothetical protein JCGZ_12380 [Jatropha curcas] |
| 8 |
Hb_001210_060 |
0.1484763031 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 9 |
Hb_000320_390 |
0.1488635184 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 10 |
Hb_101517_010 |
0.1489513844 |
- |
- |
PREDICTED: uncharacterized protein LOC105629574 [Jatropha curcas] |
| 11 |
Hb_012239_060 |
0.1493566981 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial [Jatropha curcas] |
| 12 |
Hb_189208_040 |
0.152922633 |
- |
- |
hypothetical protein POPTR_0002s18920g [Populus trichocarpa] |
| 13 |
Hb_000076_060 |
0.1541385167 |
- |
- |
hypothetical protein RCOM_0586940 [Ricinus communis] |
| 14 |
Hb_009421_070 |
0.1544262938 |
- |
- |
PREDICTED: cell number regulator 1-like [Jatropha curcas] |
| 15 |
Hb_001206_100 |
0.1557348431 |
- |
- |
hypothetical protein PRUPE_ppa021915mg, partial [Prunus persica] |
| 16 |
Hb_005842_010 |
0.1576932922 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 17 |
Hb_000329_440 |
0.1583164712 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 18 |
Hb_003835_080 |
0.1590079815 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000958_090 |
0.1592938936 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |
| 20 |
Hb_009522_020 |
0.1593130769 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |