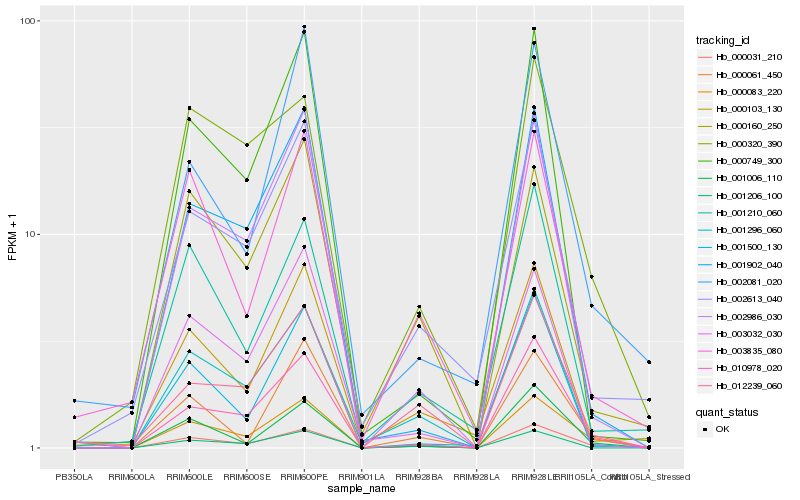
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000083_220 |
0.0 |
- |
- |
- |
| 2 |
Hb_000031_210 |
0.0478048414 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_010978_020 |
0.1109775547 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL12 [Jatropha curcas] |
| 4 |
Hb_001006_110 |
0.1275957976 |
- |
- |
PREDICTED: uncharacterized protein LOC105634212 [Jatropha curcas] |
| 5 |
Hb_002986_030 |
0.141283308 |
- |
- |
hypothetical protein JCGZ_12380 [Jatropha curcas] |
| 6 |
Hb_001296_060 |
0.1539222713 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 7 |
Hb_000061_450 |
0.1559889468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001902_040 |
0.1567198673 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000320_390 |
0.1609083627 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 10 |
Hb_000749_300 |
0.1660982036 |
- |
- |
PREDICTED: uncharacterized protein LOC105129180 [Populus euphratica] |
| 11 |
Hb_012239_060 |
0.1678992239 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000103_130 |
0.1685582831 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 13 |
Hb_001206_100 |
0.1687708002 |
- |
- |
hypothetical protein PRUPE_ppa021915mg, partial [Prunus persica] |
| 14 |
Hb_001210_060 |
0.1701773981 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 15 |
Hb_000160_250 |
0.1708114576 |
- |
- |
Uncharacterized protein TCM_019235 [Theobroma cacao] |
| 16 |
Hb_002613_040 |
0.1710149448 |
- |
- |
phosphate translocator-related family protein [Populus trichocarpa] |
| 17 |
Hb_003032_030 |
0.171050044 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002081_020 |
0.1712068629 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 19 |
Hb_001500_130 |
0.171601238 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 20 |
Hb_003835_080 |
0.1720530638 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |