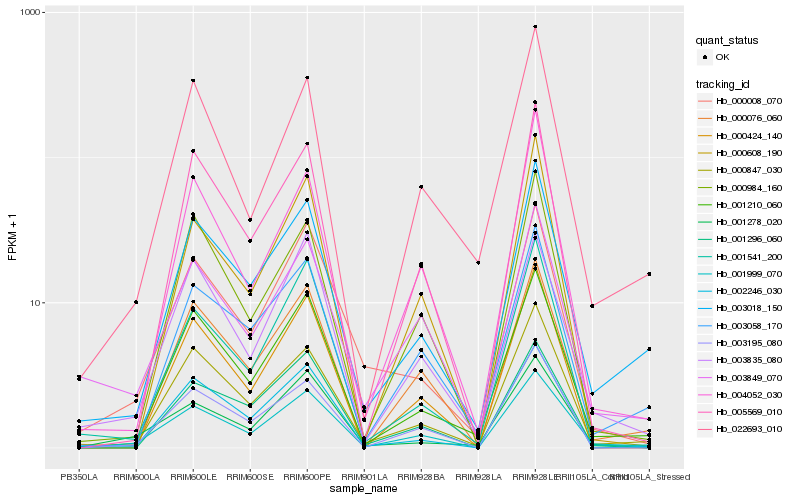
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001999_070 |
0.0 |
- |
- |
PREDICTED: ammonium transporter 1 member 3 [Jatropha curcas] |
| 2 |
Hb_003018_150 |
0.1075216761 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 3 |
Hb_001210_060 |
0.1085340237 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 4 |
Hb_000847_030 |
0.113256493 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 5 |
Hb_000076_060 |
0.1134514035 |
- |
- |
hypothetical protein RCOM_0586940 [Ricinus communis] |
| 6 |
Hb_003195_080 |
0.1147728842 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 7 |
Hb_003058_170 |
0.119987025 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003835_080 |
0.1222276457 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_003849_070 |
0.1223604775 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 10 |
Hb_000984_160 |
0.1282622464 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_005569_010 |
0.1288009191 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 12 |
Hb_001278_020 |
0.1297683631 |
- |
- |
sucrose synthase 6 [Hevea brasiliensis] |
| 13 |
Hb_000008_070 |
0.1301576976 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_022693_010 |
0.1308841579 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 15 |
Hb_001296_060 |
0.1328440491 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 16 |
Hb_002246_030 |
0.1338759993 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 17 |
Hb_000424_140 |
0.1344591667 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_001541_200 |
0.1356855348 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 19 |
Hb_004052_030 |
0.1359059892 |
- |
- |
PREDICTED: uncharacterized protein LOC105640637 [Jatropha curcas] |
| 20 |
Hb_000608_190 |
0.1379007338 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |