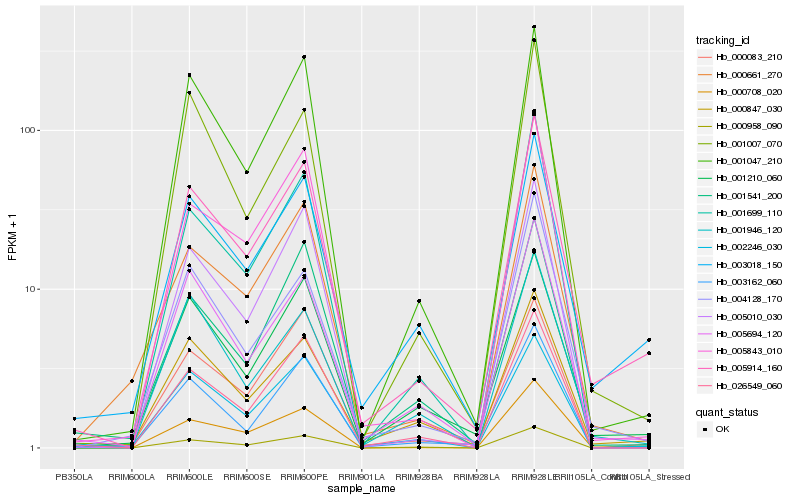
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005914_160 |
0.0 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000708_020 |
0.0817454521 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 3 |
Hb_003018_150 |
0.084830344 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 4 |
Hb_005694_120 |
0.0958875336 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 5 |
Hb_000847_030 |
0.0971018289 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 6 |
Hb_001210_060 |
0.1029461485 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 7 |
Hb_001047_210 |
0.1103305389 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 8 |
Hb_005843_010 |
0.1106468992 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 9 |
Hb_000958_090 |
0.1115655364 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |
| 10 |
Hb_002246_030 |
0.1129745929 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 11 |
Hb_004128_170 |
0.114142623 |
- |
- |
hypothetical protein POPTR_0016s08350g [Populus trichocarpa] |
| 12 |
Hb_000661_270 |
0.1151367473 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_026549_060 |
0.1157788051 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 14 |
Hb_001699_110 |
0.1199406554 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 15 |
Hb_001946_120 |
0.120595193 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 16 |
Hb_005010_030 |
0.1211748289 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 1 [Jatropha curcas] |
| 17 |
Hb_001007_070 |
0.1236487454 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 18 |
Hb_000083_210 |
0.1240496731 |
- |
- |
Major facilitator superfamily protein, putative [Theobroma cacao] |
| 19 |
Hb_003162_060 |
0.12660677 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 20 |
Hb_001541_200 |
0.1267145749 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |