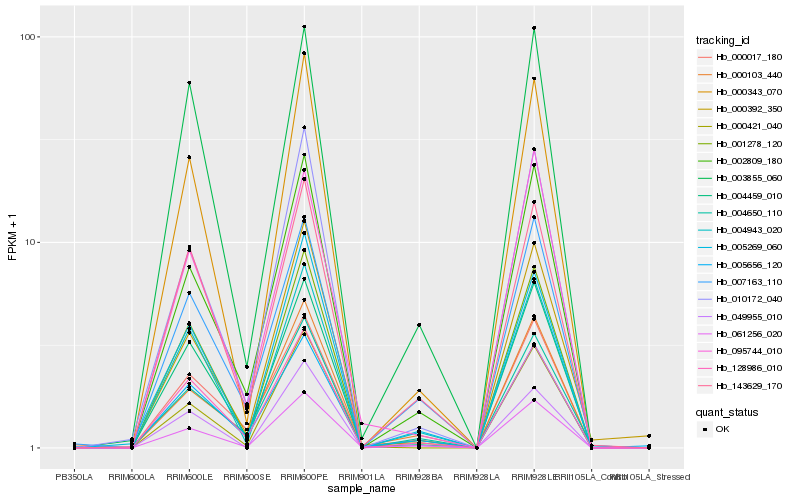
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_061256_020 |
0.0 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 2 |
Hb_000343_070 |
0.0692316665 |
- |
- |
hypothetical protein JCGZ_06265 [Jatropha curcas] |
| 3 |
Hb_002809_180 |
0.0728703417 |
- |
- |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 4 |
Hb_001278_120 |
0.0751862587 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 5 |
Hb_004943_020 |
0.0766139997 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_010172_040 |
0.0791073621 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_004459_010 |
0.0810916115 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 8 |
Hb_000103_440 |
0.0847876987 |
- |
- |
hypothetical protein POPTR_0008s15900g [Populus trichocarpa] |
| 9 |
Hb_005269_060 |
0.0866084155 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Cicer arietinum] |
| 10 |
Hb_000421_040 |
0.0873353846 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 11 |
Hb_000017_180 |
0.0911275113 |
- |
- |
hypothetical protein JCGZ_20551 [Jatropha curcas] |
| 12 |
Hb_143629_170 |
0.0941757036 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 13 |
Hb_000392_350 |
0.0979068721 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_004650_110 |
0.0986896142 |
- |
- |
hypothetical protein POPTR_0002s08760g [Populus trichocarpa] |
| 15 |
Hb_003855_060 |
0.1010698699 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 16 |
Hb_005656_120 |
0.1011224393 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 17 |
Hb_128986_010 |
0.1013925341 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007163_110 |
0.1017599686 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 19 |
Hb_049955_010 |
0.1031763376 |
- |
- |
hypothetical protein EUGRSUZ_B00742 [Eucalyptus grandis] |
| 20 |
Hb_095744_010 |
0.1036522498 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Fragaria vesca subsp. vesca] |