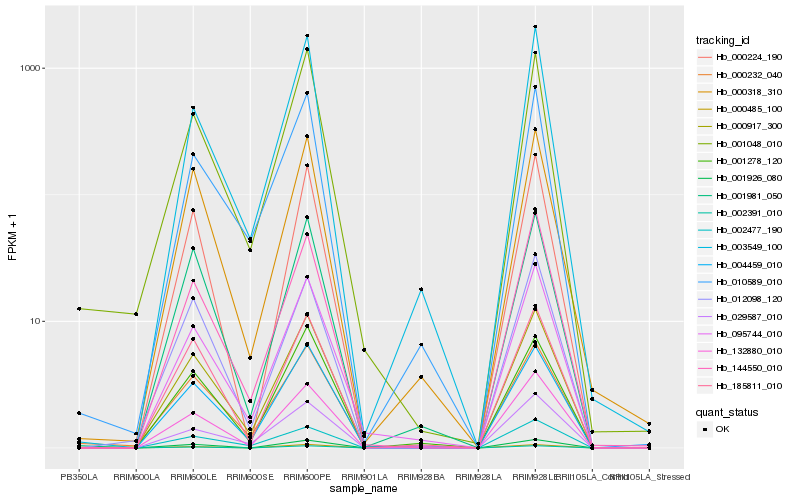
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000917_300 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Populus euphratica] |
| 2 |
Hb_004459_010 |
0.0537112248 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 3 |
Hb_001981_050 |
0.0645637534 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1-like [Jatropha curcas] |
| 4 |
Hb_000232_040 |
0.069359139 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X2 [Populus euphratica] |
| 5 |
Hb_000318_310 |
0.0697741009 |
- |
- |
hypothetical protein POPTR_0009s11190g [Populus trichocarpa] |
| 6 |
Hb_185811_010 |
0.0700094509 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 7 |
Hb_132880_010 |
0.0710975859 |
- |
- |
PREDICTED: uncharacterized protein LOC105641595 [Jatropha curcas] |
| 8 |
Hb_001048_010 |
0.0739522477 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 7 [Jatropha curcas] |
| 9 |
Hb_010589_010 |
0.074535937 |
- |
- |
PREDICTED: classical arabinogalactan protein 9-like [Jatropha curcas] |
| 10 |
Hb_095744_010 |
0.0751137429 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Fragaria vesca subsp. vesca] |
| 11 |
Hb_002477_190 |
0.0783317344 |
- |
- |
hypothetical protein EUGRSUZ_C03380, partial [Eucalyptus grandis] |
| 12 |
Hb_001926_080 |
0.0784661073 |
- |
- |
PREDICTED: uncharacterized protein LOC103454919 [Malus domestica] |
| 13 |
Hb_012098_120 |
0.0786806177 |
- |
- |
PREDICTED: putative elongation of fatty acids protein DDB_G0272012 [Jatropha curcas] |
| 14 |
Hb_029587_010 |
0.0792226936 |
- |
- |
G-type lectin S-receptor-like serine/threonine-protein kinase [Glycine soja] |
| 15 |
Hb_002391_010 |
0.0796564199 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal [Jatropha curcas] |
| 16 |
Hb_000224_190 |
0.0801253545 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_001278_120 |
0.0809014641 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 18 |
Hb_144550_010 |
0.0839820799 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 19 |
Hb_000485_100 |
0.0855715551 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Populus euphratica] |
| 20 |
Hb_003549_100 |
0.0866345069 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |