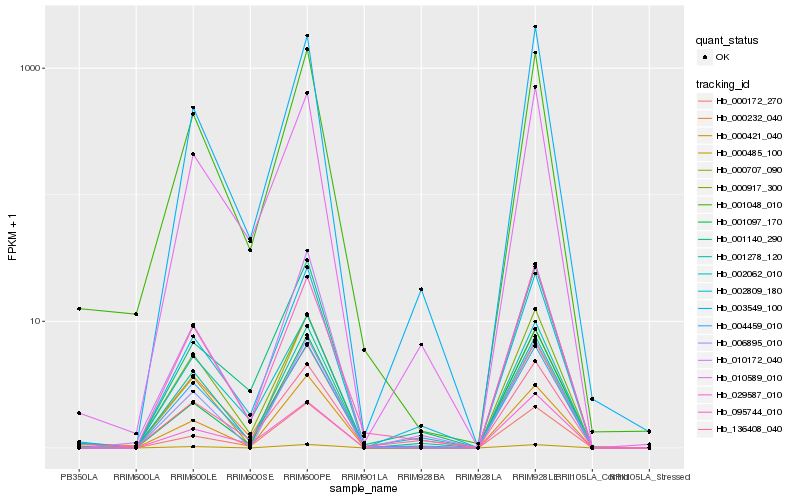
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001048_010 |
0.0 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 7 [Jatropha curcas] |
| 2 |
Hb_000917_300 |
0.0739522477 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Populus euphratica] |
| 3 |
Hb_000232_040 |
0.0756058211 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X2 [Populus euphratica] |
| 4 |
Hb_004459_010 |
0.0787314598 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 5 |
Hb_010172_040 |
0.0802512738 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000421_040 |
0.0830689667 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 7 |
Hb_010589_010 |
0.0875847243 |
- |
- |
PREDICTED: classical arabinogalactan protein 9-like [Jatropha curcas] |
| 8 |
Hb_000172_270 |
0.0891603721 |
- |
- |
hypothetical protein JCGZ_24087 [Jatropha curcas] |
| 9 |
Hb_003549_100 |
0.0933842524 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |
| 10 |
Hb_095744_010 |
0.0943989089 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Fragaria vesca subsp. vesca] |
| 11 |
Hb_002809_180 |
0.0951440462 |
- |
- |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 12 |
Hb_000707_090 |
0.0956738282 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Jatropha curcas] |
| 13 |
Hb_001278_120 |
0.0961610028 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 14 |
Hb_029587_010 |
0.0981419707 |
- |
- |
G-type lectin S-receptor-like serine/threonine-protein kinase [Glycine soja] |
| 15 |
Hb_006895_010 |
0.0987650267 |
- |
- |
PREDICTED: secologanin synthase-like [Citrus sinensis] |
| 16 |
Hb_136408_040 |
0.1001730546 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |
| 17 |
Hb_002062_010 |
0.1018749424 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 18 |
Hb_001140_290 |
0.1031807699 |
- |
- |
hypothetical protein JCGZ_26107 [Jatropha curcas] |
| 19 |
Hb_000485_100 |
0.1038465414 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Populus euphratica] |
| 20 |
Hb_001097_170 |
0.1054862686 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |