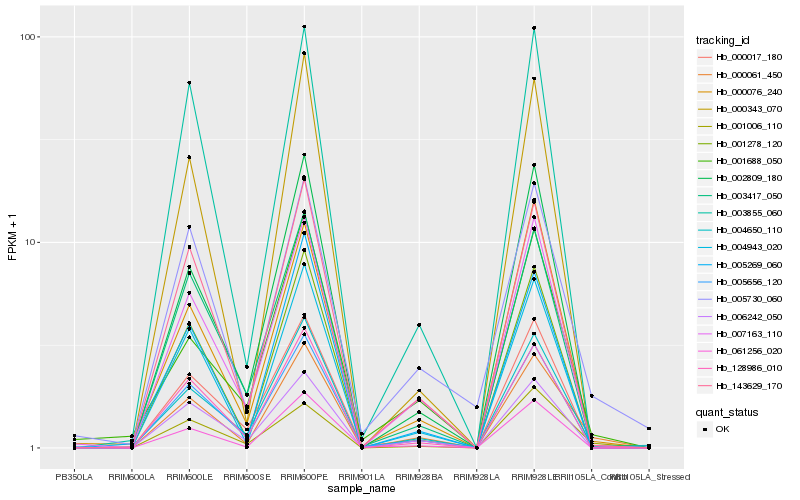
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000061_450 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004650_110 |
0.1016708412 |
- |
- |
hypothetical protein POPTR_0002s08760g [Populus trichocarpa] |
| 3 |
Hb_007163_110 |
0.1146243682 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 4 |
Hb_143629_170 |
0.1164693467 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 5 |
Hb_004943_020 |
0.1264745406 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_061256_020 |
0.131195664 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 7 |
Hb_005730_060 |
0.1316980959 |
- |
- |
PREDICTED: inorganic pyrophosphatase 1-like [Jatropha curcas] |
| 8 |
Hb_002809_180 |
0.132668605 |
- |
- |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 9 |
Hb_001278_120 |
0.1337141061 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 10 |
Hb_128986_010 |
0.1340807057 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_006242_050 |
0.1346916076 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |
| 12 |
Hb_001006_110 |
0.1348099179 |
- |
- |
PREDICTED: uncharacterized protein LOC105634212 [Jatropha curcas] |
| 13 |
Hb_001688_050 |
0.1351956105 |
- |
- |
PREDICTED: reticulon-like protein B9 [Jatropha curcas] |
| 14 |
Hb_005269_060 |
0.1385988052 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Cicer arietinum] |
| 15 |
Hb_000343_070 |
0.1393461073 |
- |
- |
hypothetical protein JCGZ_06265 [Jatropha curcas] |
| 16 |
Hb_003417_050 |
0.1395402381 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_003855_060 |
0.1397354521 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 18 |
Hb_000017_180 |
0.139900451 |
- |
- |
hypothetical protein JCGZ_20551 [Jatropha curcas] |
| 19 |
Hb_005656_120 |
0.140353505 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 20 |
Hb_000076_240 |
0.1407114882 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |