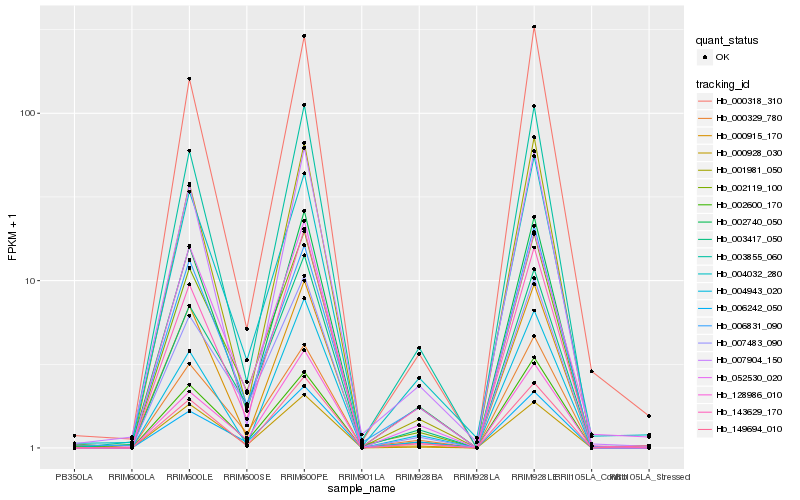
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002740_050 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86-like [Populus euphratica] |
| 2 |
Hb_003855_060 |
0.0415807593 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 3 |
Hb_007904_150 |
0.0565381896 |
- |
- |
PREDICTED: probable pectate lyase 5 [Populus euphratica] |
| 4 |
Hb_143629_170 |
0.0587018027 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 5 |
Hb_002119_100 |
0.064377443 |
- |
- |
cytochrome B561, putative [Ricinus communis] |
| 6 |
Hb_149694_010 |
0.0695639937 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_000329_780 |
0.0700922219 |
- |
- |
hypothetical protein POPTR_0403s00200g [Populus trichocarpa] |
| 8 |
Hb_001981_050 |
0.071628931 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1-like [Jatropha curcas] |
| 9 |
Hb_128986_010 |
0.0720275873 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006242_050 |
0.0741912605 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |
| 11 |
Hb_003417_050 |
0.0750570979 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_007483_090 |
0.0756066562 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004943_020 |
0.0761942279 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 14 |
Hb_000928_030 |
0.0766138535 |
transcription factor |
TF Family: bZIP |
PREDICTED: protein ABSCISIC ACID-INSENSITIVE 5 [Jatropha curcas] |
| 15 |
Hb_000915_170 |
0.0768570166 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 isoform X1 [Vitis vinifera] |
| 16 |
Hb_006831_090 |
0.0799046094 |
- |
- |
PREDICTED: uncharacterized protein LOC105642125 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004032_280 |
0.0802414824 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 18 |
Hb_000318_310 |
0.0820011282 |
- |
- |
hypothetical protein POPTR_0009s11190g [Populus trichocarpa] |
| 19 |
Hb_052530_020 |
0.0827663659 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 20 |
Hb_002600_170 |
0.0840445027 |
- |
- |
conserved hypothetical protein [Ricinus communis] |