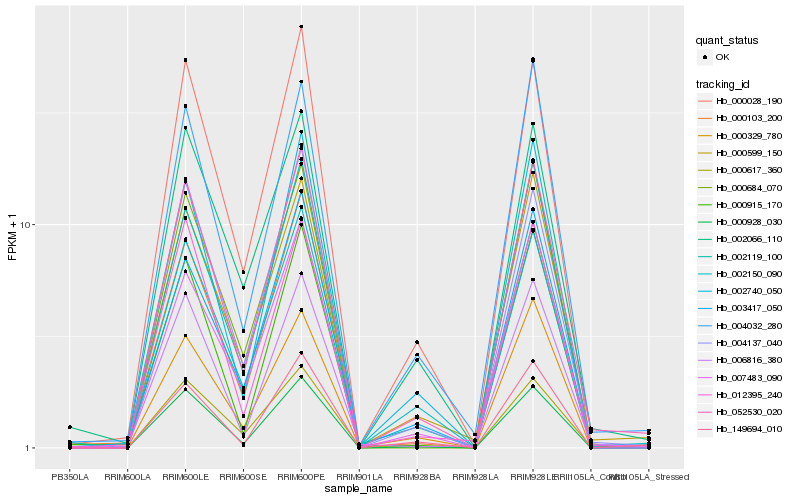
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_052530_020 |
0.0 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 2 |
Hb_000928_030 |
0.0651909273 |
transcription factor |
TF Family: bZIP |
PREDICTED: protein ABSCISIC ACID-INSENSITIVE 5 [Jatropha curcas] |
| 3 |
Hb_004137_040 |
0.0671594168 |
- |
- |
PREDICTED: serine carboxypeptidase-like 42 [Jatropha curcas] |
| 4 |
Hb_000599_150 |
0.0676675589 |
- |
- |
PREDICTED: probable arabinose 5-phosphate isomerase [Jatropha curcas] |
| 5 |
Hb_002150_090 |
0.0686690794 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 6 |
Hb_000028_190 |
0.0778900493 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 7 |
Hb_000103_200 |
0.078834859 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 8 |
Hb_003417_050 |
0.0815107965 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_002119_100 |
0.0824388878 |
- |
- |
cytochrome B561, putative [Ricinus communis] |
| 10 |
Hb_002740_050 |
0.0827663659 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86-like [Populus euphratica] |
| 11 |
Hb_006816_380 |
0.0845101204 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 12 |
Hb_149694_010 |
0.0874092632 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_000684_070 |
0.0880550836 |
- |
- |
PREDICTED: uncharacterized protein LOC105642198 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000617_360 |
0.0883315923 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004032_280 |
0.088744627 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 16 |
Hb_007483_090 |
0.0889457562 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_012395_240 |
0.0908519188 |
- |
- |
PREDICTED: uncharacterized protein LOC105638421 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000915_170 |
0.0931721855 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 isoform X1 [Vitis vinifera] |
| 19 |
Hb_002066_110 |
0.0942072682 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 20 |
Hb_000329_780 |
0.0949339769 |
- |
- |
hypothetical protein POPTR_0403s00200g [Populus trichocarpa] |