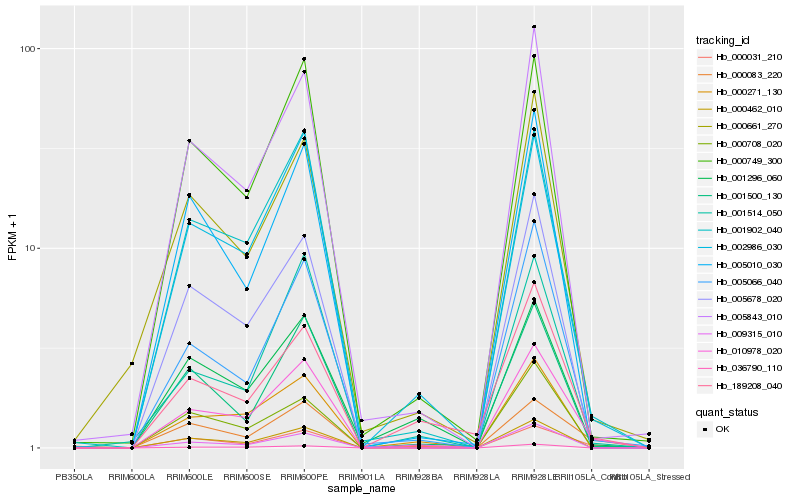
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010978_020 |
0.0 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL12 [Jatropha curcas] |
| 2 |
Hb_002986_030 |
0.0932763866 |
- |
- |
hypothetical protein JCGZ_12380 [Jatropha curcas] |
| 3 |
Hb_001902_040 |
0.0973288679 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000083_220 |
0.1109775547 |
- |
- |
- |
| 5 |
Hb_000031_210 |
0.1130585043 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_005066_040 |
0.1134747523 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 7 |
Hb_000749_300 |
0.1197111611 |
- |
- |
PREDICTED: uncharacterized protein LOC105129180 [Populus euphratica] |
| 8 |
Hb_005843_010 |
0.1229302419 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 9 |
Hb_000462_010 |
0.1265673177 |
- |
- |
hypothetical protein VITISV_041696 [Vitis vinifera] |
| 10 |
Hb_001514_050 |
0.1296133414 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 11 |
Hb_000271_130 |
0.1320574777 |
transcription factor |
TF Family: C2C2-Dof |
DNA binding with one finger 2.4, putative [Theobroma cacao] |
| 12 |
Hb_009315_010 |
0.135947087 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 13 |
Hb_005678_020 |
0.1371363952 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_001500_130 |
0.1382273313 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 15 |
Hb_000708_020 |
0.1386149475 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 16 |
Hb_000661_270 |
0.1390737418 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_036790_110 |
0.139357963 |
- |
- |
hypothetical protein JCGZ_22271 [Jatropha curcas] |
| 18 |
Hb_189208_040 |
0.1418531652 |
- |
- |
hypothetical protein POPTR_0002s18920g [Populus trichocarpa] |
| 19 |
Hb_001296_060 |
0.1420900952 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 20 |
Hb_005010_030 |
0.1421772861 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 1 [Jatropha curcas] |