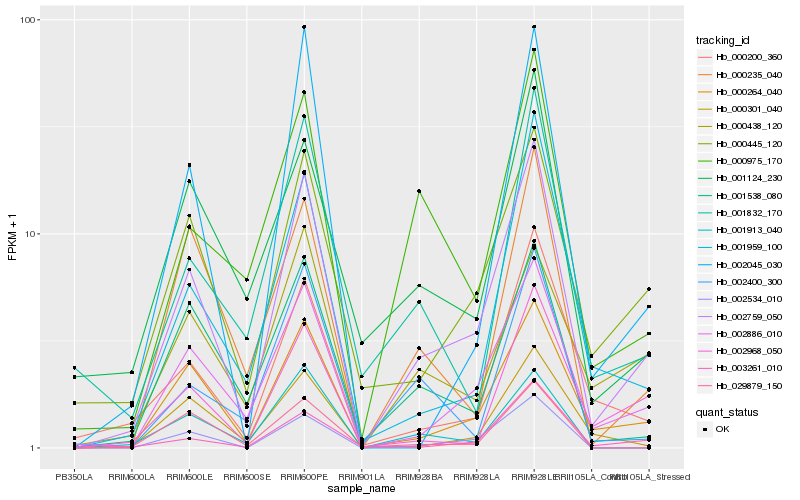
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002759_050 |
0.0 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |
| 2 |
Hb_000264_040 |
0.1205535 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 3 |
Hb_002886_010 |
0.1238835646 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000445_120 |
0.1614418901 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_003261_010 |
0.163361858 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 6 |
Hb_000200_360 |
0.1649407131 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 7 |
Hb_001959_100 |
0.165084431 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 8 |
Hb_002534_010 |
0.1664580494 |
- |
- |
PREDICTED: expansin-A1-like [Jatropha curcas] |
| 9 |
Hb_001538_080 |
0.170161981 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000235_040 |
0.1759696689 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 isoform X1 [Populus euphratica] |
| 11 |
Hb_000975_170 |
0.178212252 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 6 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002968_050 |
0.1806028068 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 13 |
Hb_002045_030 |
0.1807465625 |
transcription factor |
TF Family: AUX/IAA |
hypothetical protein CISIN_1g047687mg [Citrus sinensis] |
| 14 |
Hb_001913_040 |
0.1846537974 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000301_040 |
0.1884931119 |
- |
- |
PREDICTED: uncharacterized protein LOC105639235 [Jatropha curcas] |
| 16 |
Hb_001832_170 |
0.1886100859 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 17 |
Hb_002400_300 |
0.189325489 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 18 |
Hb_001124_230 |
0.1924456021 |
- |
- |
PREDICTED: divinyl chlorophyllide a 8-vinyl-reductase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000438_120 |
0.1952891827 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 20 |
Hb_029879_150 |
0.1963392202 |
desease resistance |
Gene Name: NB-ARC |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |