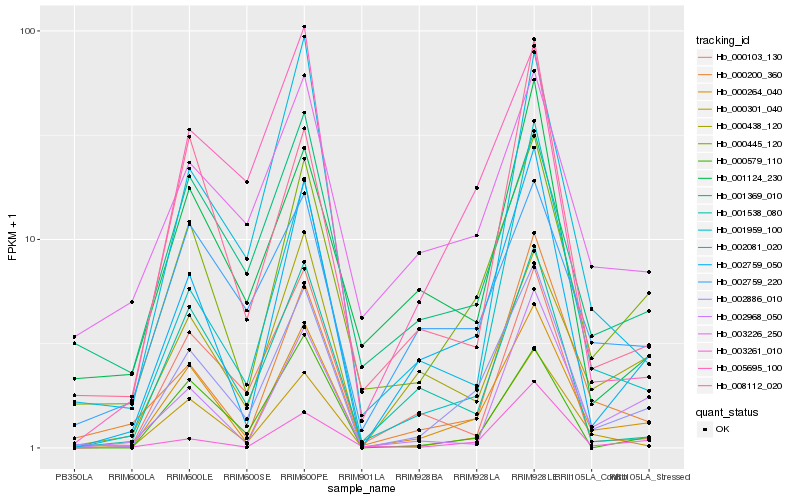
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002886_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000264_040 |
0.089123531 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 3 |
Hb_000445_120 |
0.1230764356 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_002759_050 |
0.1238835646 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |
| 5 |
Hb_000301_040 |
0.1537957574 |
- |
- |
PREDICTED: uncharacterized protein LOC105639235 [Jatropha curcas] |
| 6 |
Hb_001959_100 |
0.1738904217 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 7 |
Hb_000200_360 |
0.176486201 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 8 |
Hb_000579_110 |
0.176741027 |
- |
- |
PREDICTED: transmembrane protein 136 [Jatropha curcas] |
| 9 |
Hb_005695_100 |
0.1781918999 |
- |
- |
PREDICTED: uncharacterized protein LOC105638839 [Jatropha curcas] |
| 10 |
Hb_001538_080 |
0.1832343208 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003261_010 |
0.1867275014 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 12 |
Hb_000438_120 |
0.1871554418 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 13 |
Hb_000103_130 |
0.1929130843 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 14 |
Hb_002968_050 |
0.1959348313 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 15 |
Hb_008112_020 |
0.1964894336 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001124_230 |
0.1965963834 |
- |
- |
PREDICTED: divinyl chlorophyllide a 8-vinyl-reductase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002759_220 |
0.1969553635 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 18 |
Hb_001369_010 |
0.1974405491 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 19 |
Hb_002081_020 |
0.1976962963 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 20 |
Hb_003226_250 |
0.1978546653 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |