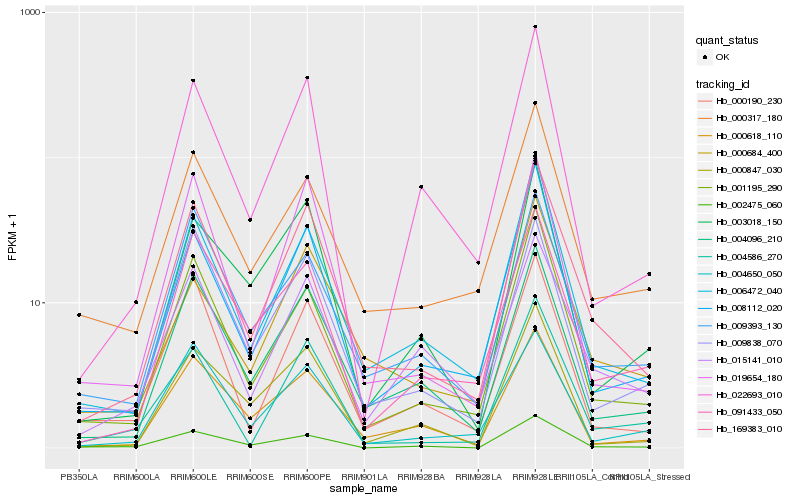
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_169383_010 |
0.0 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 2 |
Hb_006472_040 |
0.0979758056 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_004096_210 |
0.1028276805 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 4 |
Hb_008112_020 |
0.1120881347 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001195_290 |
0.1126706489 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 6 |
Hb_019654_180 |
0.1222886056 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 7 |
Hb_000618_110 |
0.127478309 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 8 |
Hb_000684_400 |
0.1276614665 |
- |
- |
PREDICTED: uncharacterized protein LOC105642159 [Jatropha curcas] |
| 9 |
Hb_000317_180 |
0.1322407236 |
- |
- |
PREDICTED: protein THYLAKOID FORMATION1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_015141_010 |
0.1324884611 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |
| 11 |
Hb_009393_130 |
0.1334645835 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 12 |
Hb_004650_050 |
0.1336678577 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_004586_270 |
0.1337813405 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000190_230 |
0.1340305457 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 15 |
Hb_009838_070 |
0.1352606236 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 16 |
Hb_091433_050 |
0.1354928421 |
- |
- |
PREDICTED: uncharacterized protein LOC105646626 [Jatropha curcas] |
| 17 |
Hb_022693_010 |
0.1389126462 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 18 |
Hb_000847_030 |
0.14185151 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 19 |
Hb_002475_060 |
0.1420314329 |
- |
- |
PREDICTED: potassium channel SKOR-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_003018_150 |
0.1422336027 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |