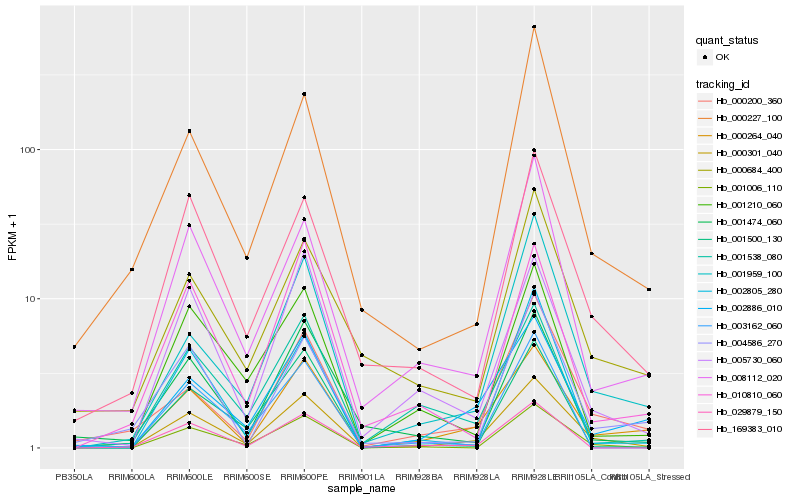
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000301_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639235 [Jatropha curcas] |
| 2 |
Hb_000264_040 |
0.1241476159 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 3 |
Hb_169383_010 |
0.1439917286 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001006_110 |
0.1443806018 |
- |
- |
PREDICTED: uncharacterized protein LOC105634212 [Jatropha curcas] |
| 5 |
Hb_001959_100 |
0.1506439727 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 6 |
Hb_002886_010 |
0.1537957574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000200_360 |
0.1578357608 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 8 |
Hb_005730_060 |
0.1588326123 |
- |
- |
PREDICTED: inorganic pyrophosphatase 1-like [Jatropha curcas] |
| 9 |
Hb_029879_150 |
0.1634992696 |
desease resistance |
Gene Name: NB-ARC |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 10 |
Hb_008112_020 |
0.1638047013 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 11 |
Hb_010810_060 |
0.171883932 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 12 |
Hb_001210_060 |
0.1722592503 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 13 |
Hb_004586_270 |
0.1725838399 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000227_100 |
0.1727372954 |
- |
- |
PREDICTED: uncharacterized protein LOC105631933 [Jatropha curcas] |
| 15 |
Hb_000684_400 |
0.1740969023 |
- |
- |
PREDICTED: uncharacterized protein LOC105642159 [Jatropha curcas] |
| 16 |
Hb_001474_060 |
0.1749841473 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 17 |
Hb_001538_080 |
0.1759285234 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002805_280 |
0.1763356912 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 19 |
Hb_003162_060 |
0.177109424 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 20 |
Hb_001500_130 |
0.1808288536 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |