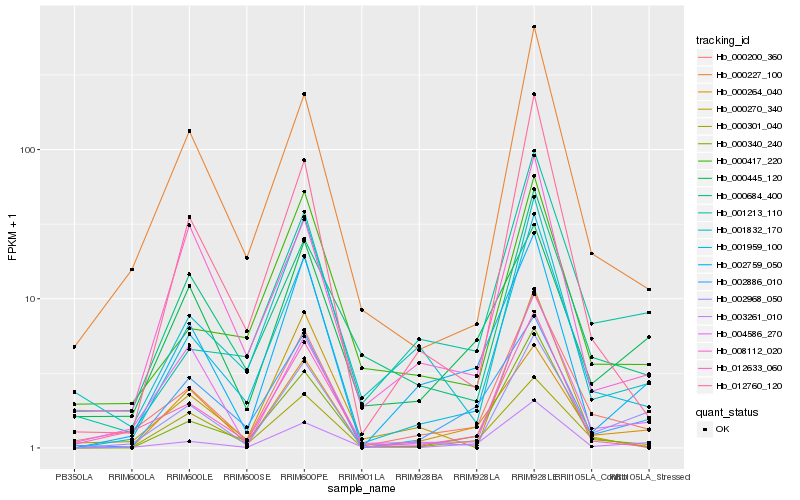
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000200_360 |
0.0 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 2 |
Hb_001959_100 |
0.111515519 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 3 |
Hb_012633_060 |
0.1316882099 |
- |
- |
laccase, putative [Ricinus communis] |
| 4 |
Hb_000227_100 |
0.1412272554 |
- |
- |
PREDICTED: uncharacterized protein LOC105631933 [Jatropha curcas] |
| 5 |
Hb_003261_010 |
0.1523838008 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 6 |
Hb_000684_400 |
0.1545215323 |
- |
- |
PREDICTED: uncharacterized protein LOC105642159 [Jatropha curcas] |
| 7 |
Hb_000340_240 |
0.155773197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000301_040 |
0.1578357608 |
- |
- |
PREDICTED: uncharacterized protein LOC105639235 [Jatropha curcas] |
| 9 |
Hb_002968_050 |
0.1579329774 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 10 |
Hb_000264_040 |
0.1647508427 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 11 |
Hb_002759_050 |
0.1649407131 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |
| 12 |
Hb_004586_270 |
0.1678148377 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000417_220 |
0.1693427804 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Cucumis melo] |
| 14 |
Hb_001832_170 |
0.1715495745 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 15 |
Hb_012760_120 |
0.1736152155 |
- |
- |
PREDICTED: peroxidase 64-like [Jatropha curcas] |
| 16 |
Hb_002886_010 |
0.176486201 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001213_110 |
0.1781916794 |
- |
- |
SOUL heme-binding family protein [Populus trichocarpa] |
| 18 |
Hb_008112_020 |
0.1807791601 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000445_120 |
0.182671684 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_000270_340 |
0.1849497465 |
- |
- |
hypothetical protein JCGZ_11546 [Jatropha curcas] |