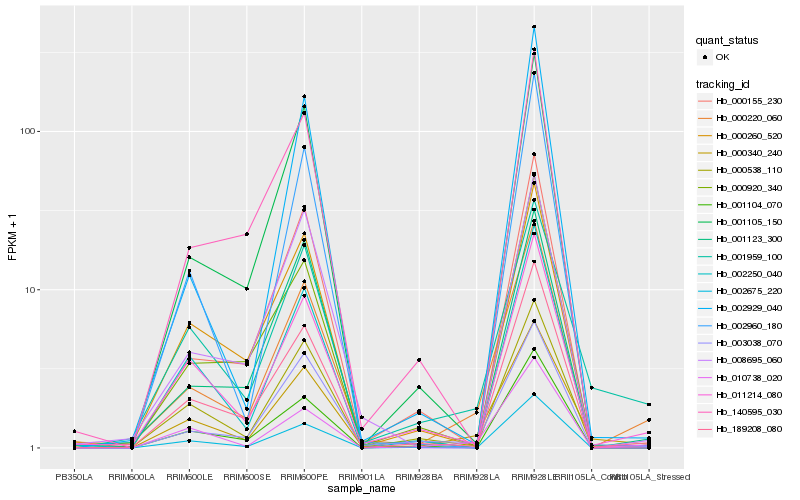
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000220_060 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 15 [Jatropha curcas] |
| 2 |
Hb_189208_080 |
0.1106326833 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 11-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_003038_070 |
0.1131959304 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 4 |
Hb_000340_240 |
0.1146330209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_011214_080 |
0.1217808703 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 6 |
Hb_010738_020 |
0.1240680767 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 7 |
Hb_001105_150 |
0.1352178964 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |
| 8 |
Hb_000155_230 |
0.1384166735 |
- |
- |
rac gtpase, putative [Ricinus communis] |
| 9 |
Hb_000538_110 |
0.1403350886 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 10 |
Hb_008695_060 |
0.1437984503 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 11 |
Hb_002675_220 |
0.1454522208 |
- |
- |
PREDICTED: probable pyridoxal biosynthesis protein PDX1.2 [Populus euphratica] |
| 12 |
Hb_001959_100 |
0.1470366544 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 13 |
Hb_001123_300 |
0.1482600898 |
- |
- |
hypothetical protein POPTR_0003s16270g [Populus trichocarpa] |
| 14 |
Hb_002960_180 |
0.1488949888 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 15 |
Hb_000920_340 |
0.1496111601 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_001104_070 |
0.1500496946 |
- |
- |
hypothetical protein SORBIDRAFT_10g024640 [Sorghum bicolor] |
| 17 |
Hb_140595_030 |
0.1513350289 |
- |
- |
- |
| 18 |
Hb_002250_040 |
0.1517017643 |
- |
- |
PREDICTED: transcription repressor OFP17 [Jatropha curcas] |
| 19 |
Hb_002929_040 |
0.1523521604 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_000260_520 |
0.1537103482 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |