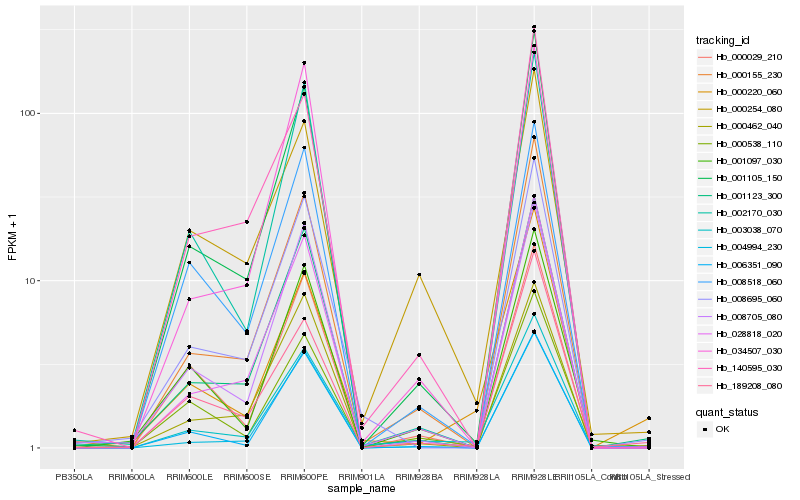
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003038_070 |
0.0 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 2 |
Hb_000155_230 |
0.0965020067 |
- |
- |
rac gtpase, putative [Ricinus communis] |
| 3 |
Hb_028818_020 |
0.0972394993 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9-like [Jatropha curcas] |
| 4 |
Hb_001123_300 |
0.1018338376 |
- |
- |
hypothetical protein POPTR_0003s16270g [Populus trichocarpa] |
| 5 |
Hb_001105_150 |
0.1033294957 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |
| 6 |
Hb_000538_110 |
0.1065838339 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 7 |
Hb_008695_060 |
0.110737854 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 8 |
Hb_000220_060 |
0.1131959304 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 15 [Jatropha curcas] |
| 9 |
Hb_000254_080 |
0.1187282892 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 10 |
Hb_002170_030 |
0.1217966587 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 11 |
Hb_008705_080 |
0.1248234177 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 12 |
Hb_006351_090 |
0.1251493462 |
- |
- |
UDP-glycosyltransferase 85A1 [Gossypium arboreum] |
| 13 |
Hb_004994_230 |
0.1258606696 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3, putative [Ricinus communis] |
| 14 |
Hb_189208_080 |
0.1272404367 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 11-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_140595_030 |
0.1274003722 |
- |
- |
- |
| 16 |
Hb_034507_030 |
0.1276112455 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 17 |
Hb_001097_030 |
0.1280042959 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 18 |
Hb_008518_060 |
0.1280112418 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 1 [Jatropha curcas] |
| 19 |
Hb_000462_040 |
0.1289905748 |
- |
- |
ATSMC2, putative [Ricinus communis] |
| 20 |
Hb_000029_210 |
0.1291040767 |
- |
- |
protein binding protein, putative [Ricinus communis] |