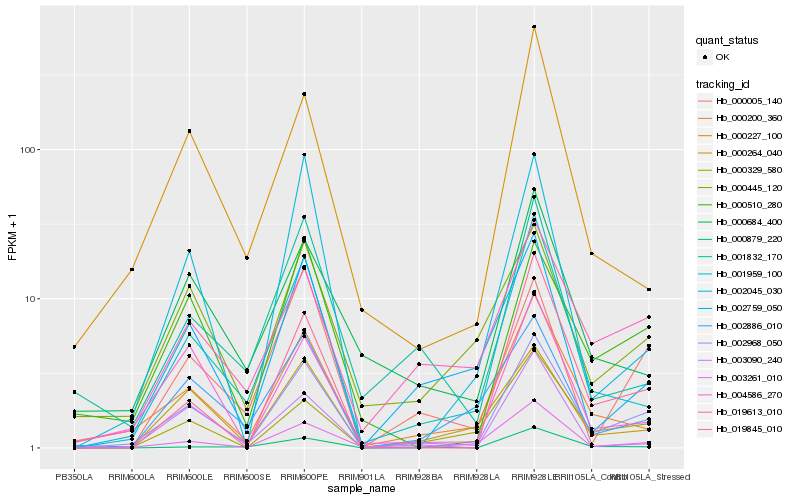
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002968_050 |
0.0 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 2 |
Hb_003261_010 |
0.1576342754 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 3 |
Hb_000200_360 |
0.1579329774 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 4 |
Hb_001959_100 |
0.1585217839 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 5 |
Hb_000510_280 |
0.16947989 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like [Jatropha curcas] |
| 6 |
Hb_000264_040 |
0.1706391941 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 7 |
Hb_004586_270 |
0.1752546804 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_019613_010 |
0.1763744635 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_000329_580 |
0.1797346553 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_002759_050 |
0.1806028068 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |
| 11 |
Hb_019845_010 |
0.1861558293 |
- |
- |
protease inhibitor protein [Hevea brasiliensis] |
| 12 |
Hb_000445_120 |
0.1890435011 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_000005_140 |
0.1958506094 |
transcription factor |
TF Family: C2H2 |
unknown [Populus trichocarpa] |
| 14 |
Hb_002886_010 |
0.1959348313 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000227_100 |
0.1963009442 |
- |
- |
PREDICTED: uncharacterized protein LOC105631933 [Jatropha curcas] |
| 16 |
Hb_000684_400 |
0.1982040022 |
- |
- |
PREDICTED: uncharacterized protein LOC105642159 [Jatropha curcas] |
| 17 |
Hb_002045_030 |
0.2016061455 |
transcription factor |
TF Family: AUX/IAA |
hypothetical protein CISIN_1g047687mg [Citrus sinensis] |
| 18 |
Hb_003090_240 |
0.20335232 |
- |
- |
hypothetical protein JCGZ_25095 [Jatropha curcas] |
| 19 |
Hb_000879_220 |
0.2096354897 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 20 |
Hb_001832_170 |
0.2096492199 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |