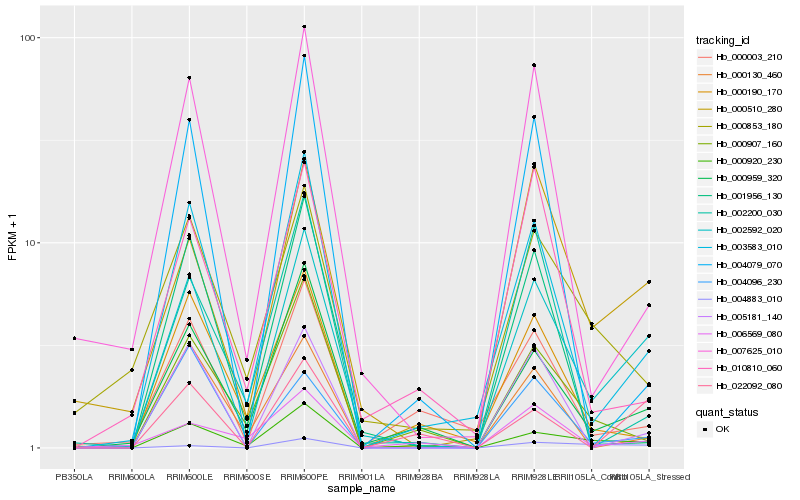
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002592_020 |
0.0 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003583_010 |
0.1670200904 |
- |
- |
PREDICTED: patatin-like protein 6 [Jatropha curcas] |
| 3 |
Hb_000510_280 |
0.1744665404 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like [Jatropha curcas] |
| 4 |
Hb_000959_320 |
0.1746460189 |
- |
- |
PREDICTED: uncharacterized protein LOC105778325 [Gossypium raimondii] |
| 5 |
Hb_000920_230 |
0.1938482945 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_005181_140 |
0.2051196873 |
- |
- |
hypothetical protein JCGZ_19000 [Jatropha curcas] |
| 7 |
Hb_007625_010 |
0.2068484489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000003_210 |
0.22665306 |
- |
- |
PREDICTED: GEM-like protein 4 [Jatropha curcas] |
| 9 |
Hb_022092_080 |
0.2281784202 |
- |
- |
- |
| 10 |
Hb_000130_460 |
0.2295745199 |
- |
- |
hypothetical protein JCGZ_15296 [Jatropha curcas] |
| 11 |
Hb_000853_180 |
0.2302725218 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 12 |
Hb_004079_070 |
0.2391883017 |
- |
- |
hypothetical protein JCGZ_07230 [Jatropha curcas] |
| 13 |
Hb_000907_160 |
0.2406346142 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 14 |
Hb_010810_060 |
0.2417112287 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 15 |
Hb_000190_170 |
0.2417609442 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 16 |
Hb_001956_130 |
0.241803609 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_004096_230 |
0.2419933582 |
- |
- |
PREDICTED: uncharacterized protein LOC105638704 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006569_080 |
0.24292173 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_024559 [Vitis vinifera] |
| 19 |
Hb_002200_030 |
0.2440383826 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 20 |
Hb_004883_010 |
0.2442809468 |
- |
- |
hypothetical protein JCGZ_23144 [Jatropha curcas] |