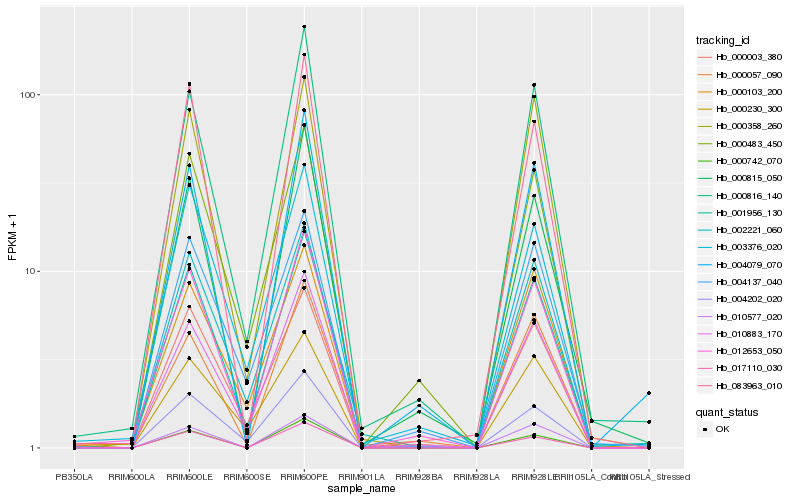
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001956_130 |
0.0 |
- |
- |
catalytic, putative [Ricinus communis] |
| 2 |
Hb_002221_060 |
0.0770362606 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 3 |
Hb_000816_140 |
0.0838960084 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 4 |
Hb_000003_380 |
0.0883295316 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_010883_170 |
0.0907441069 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 6 |
Hb_012653_050 |
0.0939866416 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 7 |
Hb_000358_260 |
0.0946102208 |
- |
- |
PREDICTED: lachrymatory-factor synthase-like [Vitis vinifera] |
| 8 |
Hb_003376_020 |
0.0965553699 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000483_450 |
0.0969035097 |
- |
- |
PREDICTED: anthranilate N-benzoyltransferase protein 1 [Jatropha curcas] |
| 10 |
Hb_083963_010 |
0.0987835889 |
- |
- |
PREDICTED: probable carboxylesterase 8 [Jatropha curcas] |
| 11 |
Hb_004137_040 |
0.100417355 |
- |
- |
PREDICTED: serine carboxypeptidase-like 42 [Jatropha curcas] |
| 12 |
Hb_000815_050 |
0.1009650941 |
- |
- |
PREDICTED: expansin-A1 [Populus euphratica] |
| 13 |
Hb_000742_070 |
0.101589788 |
- |
- |
PREDICTED: monoglyceride lipase isoform X2 [Bubalus bubalis] |
| 14 |
Hb_017110_030 |
0.1035078104 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.4 [Jatropha curcas] |
| 15 |
Hb_004202_020 |
0.1047914056 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000103_200 |
0.1067836689 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 17 |
Hb_010577_020 |
0.1083672203 |
- |
- |
- |
| 18 |
Hb_000230_300 |
0.109647671 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme 4 [Jatropha curcas] |
| 19 |
Hb_004079_070 |
0.1098567135 |
- |
- |
hypothetical protein JCGZ_07230 [Jatropha curcas] |
| 20 |
Hb_000057_090 |
0.1108001427 |
- |
- |
hypothetical protein RCOM_1494490 [Ricinus communis] |