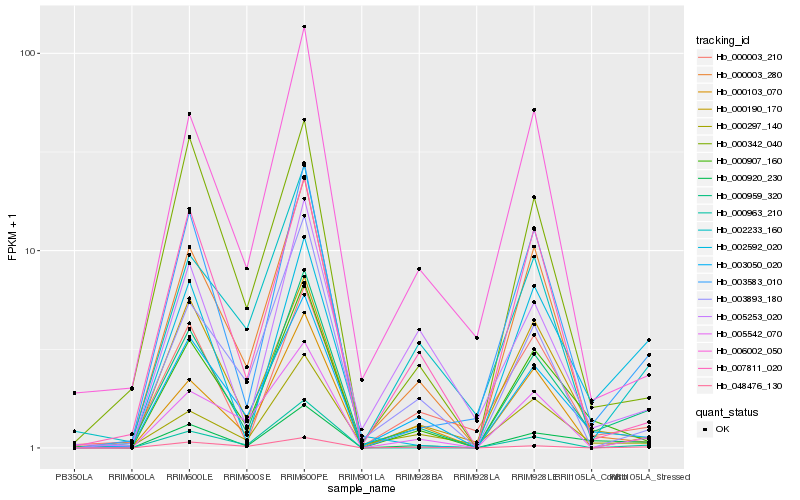
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000959_320 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105778325 [Gossypium raimondii] |
| 2 |
Hb_003050_020 |
0.1255283835 |
- |
- |
glycosyl hydrolase family 17 family protein [Populus trichocarpa] |
| 3 |
Hb_003583_010 |
0.1290192727 |
- |
- |
PREDICTED: patatin-like protein 6 [Jatropha curcas] |
| 4 |
Hb_000907_160 |
0.1404990183 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 5 |
Hb_000920_230 |
0.1458168969 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000103_070 |
0.1475869457 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 7 |
Hb_000003_210 |
0.1585481824 |
- |
- |
PREDICTED: GEM-like protein 4 [Jatropha curcas] |
| 8 |
Hb_003893_180 |
0.1612535937 |
- |
- |
PREDICTED: uncharacterized protein LOC105648771 [Jatropha curcas] |
| 9 |
Hb_006002_050 |
0.1655859324 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 10 |
Hb_000297_140 |
0.1670051604 |
- |
- |
hypothetical protein RCOM_0293450 [Ricinus communis] |
| 11 |
Hb_000003_280 |
0.1681853681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000342_040 |
0.1726836755 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 13 |
Hb_007811_020 |
0.1735682246 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 14 |
Hb_000963_210 |
0.1737254386 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 15 |
Hb_005542_070 |
0.1744468309 |
- |
- |
hypothetical protein ZEAMMB73_851878 [Zea mays] |
| 16 |
Hb_002233_160 |
0.1745192098 |
- |
- |
PREDICTED: uncharacterized protein LOC105633094 [Jatropha curcas] |
| 17 |
Hb_002592_020 |
0.1746460189 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000190_170 |
0.1750614481 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 19 |
Hb_005253_020 |
0.1759252924 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 20 |
Hb_048476_130 |
0.1783910937 |
- |
- |
scab resistance family protein [Populus trichocarpa] |