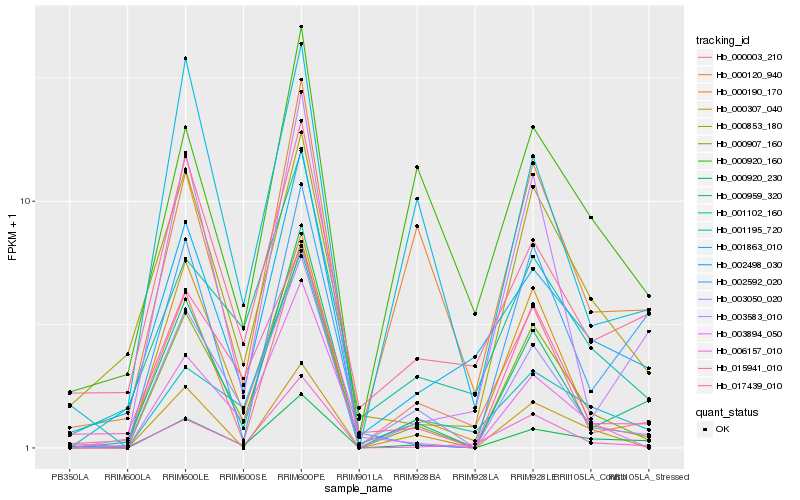
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000920_230 |
0.0 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_000959_320 |
0.1458168969 |
- |
- |
PREDICTED: uncharacterized protein LOC105778325 [Gossypium raimondii] |
| 3 |
Hb_000907_160 |
0.1520396147 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 4 |
Hb_002498_030 |
0.1748537817 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003050_020 |
0.1783993524 |
- |
- |
glycosyl hydrolase family 17 family protein [Populus trichocarpa] |
| 6 |
Hb_017439_010 |
0.1824566304 |
- |
- |
putative actin-97 protein [Linum usitatissimum] |
| 7 |
Hb_002592_020 |
0.1938482945 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000120_940 |
0.1984498299 |
- |
- |
PREDICTED: phosphoserine aminotransferase 1, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000853_180 |
0.1986653165 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 10 |
Hb_003583_010 |
0.2009033124 |
- |
- |
PREDICTED: patatin-like protein 6 [Jatropha curcas] |
| 11 |
Hb_000003_210 |
0.2064701404 |
- |
- |
PREDICTED: GEM-like protein 4 [Jatropha curcas] |
| 12 |
Hb_006157_010 |
0.2083446383 |
- |
- |
PREDICTED: endoglucanase 5 [Jatropha curcas] |
| 13 |
Hb_000190_170 |
0.210419347 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 14 |
Hb_015941_010 |
0.2114013725 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 15 |
Hb_000307_040 |
0.2138159467 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 16 |
Hb_001863_010 |
0.2175810347 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 17 |
Hb_001102_160 |
0.2186129871 |
- |
- |
PREDICTED: galacturonokinase [Jatropha curcas] |
| 18 |
Hb_000920_160 |
0.2196599102 |
- |
- |
annexin [Manihot esculenta] |
| 19 |
Hb_003894_050 |
0.2224328768 |
- |
- |
hypothetical protein JCGZ_14524 [Jatropha curcas] |
| 20 |
Hb_001195_720 |
0.2241214014 |
- |
- |
PREDICTED: probable calcium-binding protein CML41 [Jatropha curcas] |