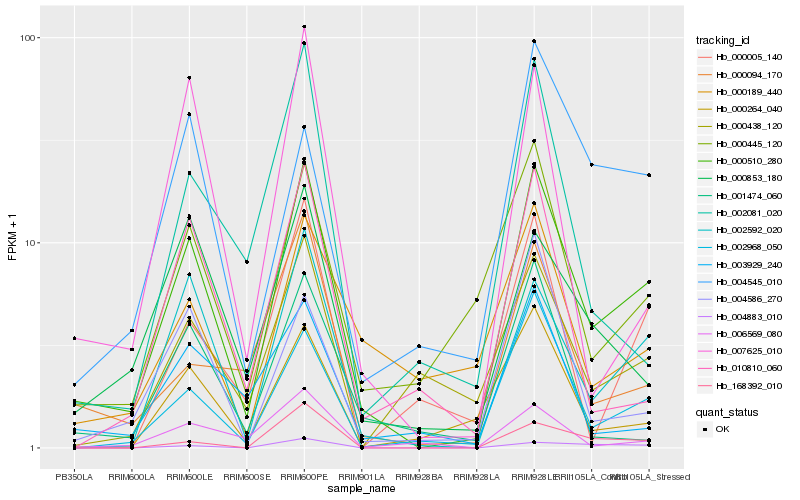
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000510_280 |
0.0 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like [Jatropha curcas] |
| 2 |
Hb_002968_050 |
0.16947989 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 3 |
Hb_002592_020 |
0.1744665404 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000445_120 |
0.1926521485 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_000264_040 |
0.2055612865 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 6 |
Hb_000094_170 |
0.2094064656 |
- |
- |
PREDICTED: uncharacterized protein LOC105633781 [Jatropha curcas] |
| 7 |
Hb_000438_120 |
0.2159286494 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 8 |
Hb_007625_010 |
0.2170452962 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004586_270 |
0.2178327681 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_006569_080 |
0.2185877485 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_024559 [Vitis vinifera] |
| 11 |
Hb_010810_060 |
0.2197240181 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 12 |
Hb_003929_240 |
0.2197279509 |
- |
- |
PREDICTED: uncharacterized protein LOC105643616 isoform X2 [Jatropha curcas] |
| 13 |
Hb_168392_010 |
0.2227524081 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 14 |
Hb_000853_180 |
0.2236268133 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 15 |
Hb_001474_060 |
0.2260372512 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 16 |
Hb_000005_140 |
0.2260760918 |
transcription factor |
TF Family: C2H2 |
unknown [Populus trichocarpa] |
| 17 |
Hb_002081_020 |
0.2263397898 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 18 |
Hb_004883_010 |
0.2276059612 |
- |
- |
hypothetical protein JCGZ_23144 [Jatropha curcas] |
| 19 |
Hb_004545_010 |
0.2284123177 |
- |
- |
acyl carrier protein [Ricinus communis] |
| 20 |
Hb_000189_440 |
0.2291253364 |
- |
- |
hypothetical protein JCGZ_23470 [Jatropha curcas] |