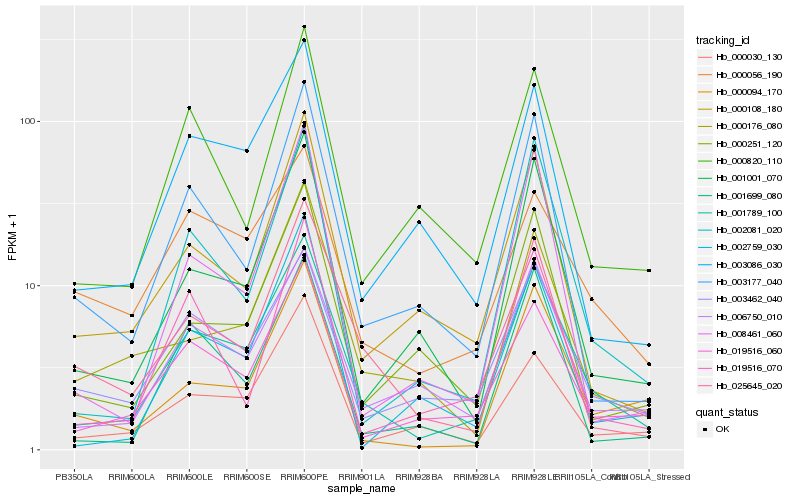
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001789_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_019516_060 |
0.1210906905 |
- |
- |
PREDICTED: uncharacterized protein LOC105634259 [Jatropha curcas] |
| 3 |
Hb_006750_010 |
0.1261544212 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 4 |
Hb_003462_040 |
0.136758431 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_001001_070 |
0.146978245 |
- |
- |
PREDICTED: uncharacterized protein LOC105633663 [Jatropha curcas] |
| 6 |
Hb_003086_030 |
0.1472606923 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 7 |
Hb_025645_020 |
0.1491141962 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 8 |
Hb_002081_020 |
0.1492152228 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 9 |
Hb_000108_180 |
0.1529868992 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 10 |
Hb_003177_040 |
0.1532265233 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 11 |
Hb_000094_170 |
0.1553256151 |
- |
- |
PREDICTED: uncharacterized protein LOC105633781 [Jatropha curcas] |
| 12 |
Hb_000030_130 |
0.1556418988 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000176_080 |
0.1578314727 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g30520 [Jatropha curcas] |
| 14 |
Hb_000251_120 |
0.1594135242 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 15 |
Hb_001699_080 |
0.1596902981 |
- |
- |
unnamed protein product [Coffea canephora] |
| 16 |
Hb_000820_110 |
0.16289297 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 17 |
Hb_000056_190 |
0.1649576357 |
- |
- |
hypothetical protein [Ricinus communis] |
| 18 |
Hb_019516_070 |
0.1695226643 |
- |
- |
PREDICTED: uncharacterized protein LOC105634258 [Jatropha curcas] |
| 19 |
Hb_002759_030 |
0.1701531042 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 20 |
Hb_008461_060 |
0.1713944314 |
- |
- |
hypothetical protein JCGZ_17379 [Jatropha curcas] |