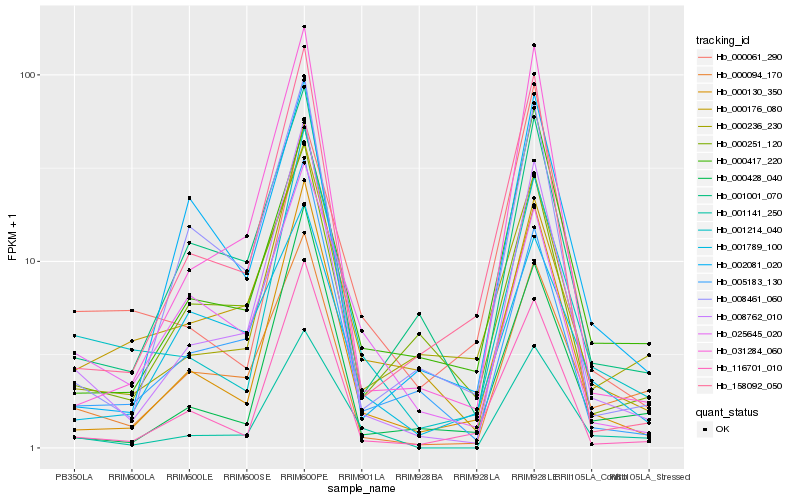
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001141_250 |
0.0 |
- |
- |
hypothetical protein glysoja_047929, partial [Glycine soja] |
| 2 |
Hb_001214_040 |
0.1421877911 |
- |
- |
PREDICTED: protein trichome birefringence-like 3 [Jatropha curcas] |
| 3 |
Hb_008762_010 |
0.1490671635 |
- |
- |
dopamine beta-monooxygenase, putative [Ricinus communis] |
| 4 |
Hb_000094_170 |
0.1565057034 |
- |
- |
PREDICTED: uncharacterized protein LOC105633781 [Jatropha curcas] |
| 5 |
Hb_000130_350 |
0.1571650069 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 6 |
Hb_000417_220 |
0.1741370635 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Cucumis melo] |
| 7 |
Hb_000428_040 |
0.1904687326 |
- |
- |
PREDICTED: protein trichome birefringence-like 31 [Populus euphratica] |
| 8 |
Hb_000236_230 |
0.1905398339 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At4g11680 isoform X2 [Jatropha curcas] |
| 9 |
Hb_116701_010 |
0.1908121267 |
- |
- |
hypothetical protein POPTR_0005s19410g [Populus trichocarpa] |
| 10 |
Hb_031284_060 |
0.1920464894 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 7 [UDP-forming] [Jatropha curcas] |
| 11 |
Hb_025645_020 |
0.1959229976 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 12 |
Hb_000176_080 |
0.1976058442 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g30520 [Jatropha curcas] |
| 13 |
Hb_001001_070 |
0.1999831598 |
- |
- |
PREDICTED: uncharacterized protein LOC105633663 [Jatropha curcas] |
| 14 |
Hb_001789_100 |
0.2116602994 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000251_120 |
0.2134056051 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 16 |
Hb_008461_060 |
0.2136096708 |
- |
- |
hypothetical protein JCGZ_17379 [Jatropha curcas] |
| 17 |
Hb_158092_050 |
0.2139843879 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10 [Jatropha curcas] |
| 18 |
Hb_005183_130 |
0.2147989451 |
- |
- |
glycogenin, putative [Ricinus communis] |
| 19 |
Hb_000061_290 |
0.2165151431 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002081_020 |
0.2187619087 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |