| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
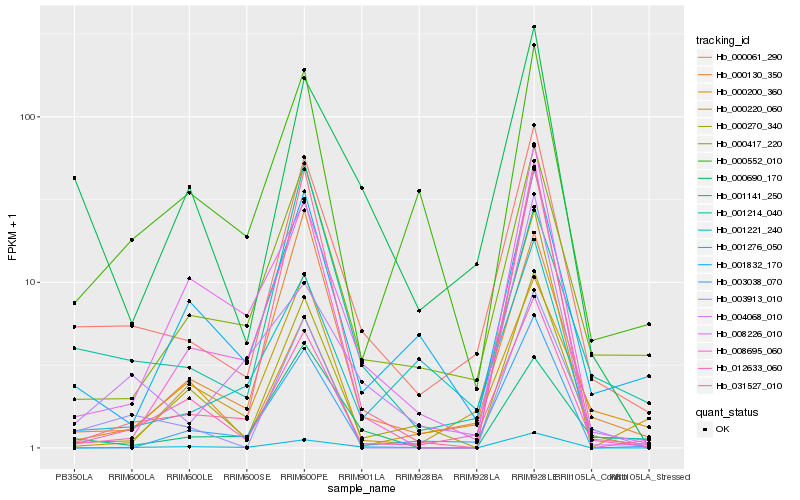
Hb_000061_290 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000417_220 |
0.1538430566 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Cucumis melo] |
| 3 |
Hb_012633_060 |
0.1573400041 |
- |
- |
laccase, putative [Ricinus communis] |
| 4 |
Hb_000690_170 |
0.1657923812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003913_010 |
0.1834038346 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001276_050 |
0.1885463589 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance-like protein 1O [Medicago truncatula] |
| 7 |
Hb_000270_340 |
0.1919042789 |
- |
- |
hypothetical protein JCGZ_11546 [Jatropha curcas] |
| 8 |
Hb_001832_170 |
0.1922990238 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 9 |
Hb_000130_350 |
0.1979404273 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 10 |
Hb_001221_240 |
0.1979926394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003038_070 |
0.2029598645 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 12 |
Hb_000200_360 |
0.2039182734 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 13 |
Hb_000552_010 |
0.2077485333 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 2 [Jatropha curcas] |
| 14 |
Hb_000220_060 |
0.2142215621 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 15 [Jatropha curcas] |
| 15 |
Hb_008695_060 |
0.2160075767 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 16 |
Hb_004068_010 |
0.2161144532 |
- |
- |
beta-amyrin synthase [Euphorbia tirucalli] |
| 17 |
Hb_001141_250 |
0.2165151431 |
- |
- |
hypothetical protein glysoja_047929, partial [Glycine soja] |
| 18 |
Hb_008226_010 |
0.216860658 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 19 |
Hb_001214_040 |
0.2172176139 |
- |
- |
PREDICTED: protein trichome birefringence-like 3 [Jatropha curcas] |
| 20 |
Hb_031527_010 |
0.2173147814 |
- |
- |
conserved hypothetical protein [Ricinus communis] |