| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
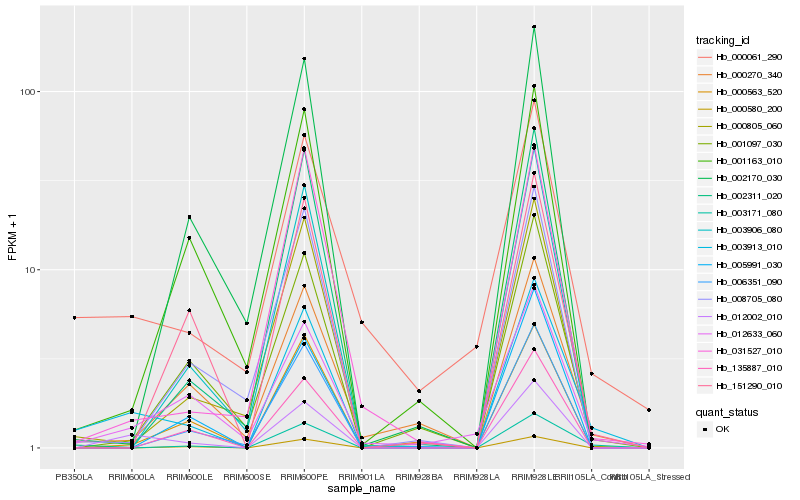
Hb_003913_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_012002_010 |
0.1527333347 |
- |
- |
hypothetical protein VITISV_016116 [Vitis vinifera] |
| 3 |
Hb_003171_080 |
0.1761539565 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000061_290 |
0.1834038346 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_012633_060 |
0.1860042613 |
- |
- |
laccase, putative [Ricinus communis] |
| 6 |
Hb_000805_060 |
0.1922323909 |
- |
- |
laccase, putative [Ricinus communis] |
| 7 |
Hb_000270_340 |
0.1987277101 |
- |
- |
hypothetical protein JCGZ_11546 [Jatropha curcas] |
| 8 |
Hb_003906_080 |
0.2044696738 |
- |
- |
Peroxidase 66 precursor, putative [Ricinus communis] |
| 9 |
Hb_000563_520 |
0.206038254 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 10 |
Hb_002311_020 |
0.2061774149 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 11 |
Hb_001097_030 |
0.2082240144 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 12 |
Hb_008705_080 |
0.2091842594 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 13 |
Hb_031527_010 |
0.2096001263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006351_090 |
0.210852634 |
- |
- |
UDP-glycosyltransferase 85A1 [Gossypium arboreum] |
| 15 |
Hb_000580_200 |
0.2129069599 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001163_010 |
0.2144719078 |
- |
- |
PREDICTED: syncoilin [Jatropha curcas] |
| 17 |
Hb_002170_030 |
0.2147708714 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 18 |
Hb_151290_010 |
0.2148912709 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_135887_010 |
0.215440863 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 20 |
Hb_005991_030 |
0.2174100105 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |