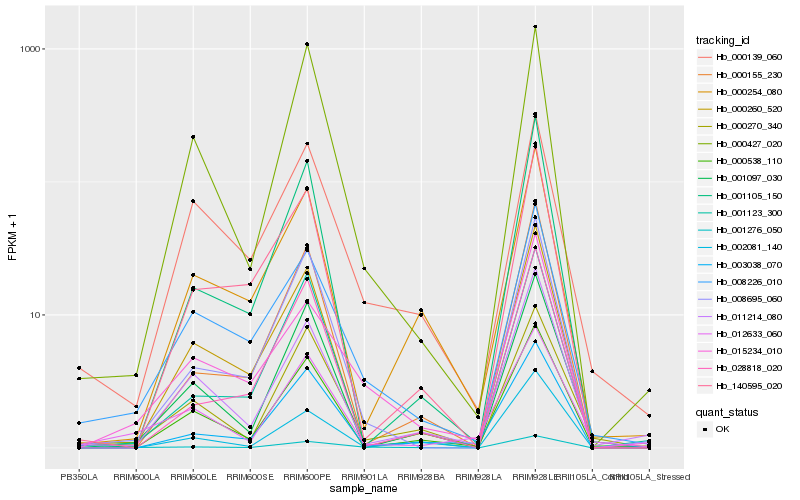
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001276_050 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance-like protein 1O [Medicago truncatula] |
| 2 |
Hb_015234_010 |
0.127541436 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 4B [Jatropha curcas] |
| 3 |
Hb_008226_010 |
0.1370277242 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 4 |
Hb_000270_340 |
0.1536527667 |
- |
- |
hypothetical protein JCGZ_11546 [Jatropha curcas] |
| 5 |
Hb_008695_060 |
0.1551976195 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 6 |
Hb_003038_070 |
0.1650050694 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 7 |
Hb_001097_030 |
0.1675476517 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 8 |
Hb_000427_020 |
0.171754418 |
- |
- |
PsbA, partial [Dianthus pygmaeus] |
| 9 |
Hb_000139_060 |
0.1720221561 |
- |
- |
Photosystem I reaction center subunit II, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_000538_110 |
0.1724524932 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 11 |
Hb_000155_230 |
0.1725942131 |
- |
- |
rac gtpase, putative [Ricinus communis] |
| 12 |
Hb_000254_080 |
0.1737043921 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 13 |
Hb_002081_140 |
0.1744180836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001123_300 |
0.1760989411 |
- |
- |
hypothetical protein POPTR_0003s16270g [Populus trichocarpa] |
| 15 |
Hb_000260_520 |
0.1761406979 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 16 |
Hb_001105_150 |
0.1765036972 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |
| 17 |
Hb_028818_020 |
0.1770440332 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9-like [Jatropha curcas] |
| 18 |
Hb_012633_060 |
0.1783967857 |
- |
- |
laccase, putative [Ricinus communis] |
| 19 |
Hb_011214_080 |
0.1808976233 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 20 |
Hb_140595_020 |
0.1809074771 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Solanum lycopersicum] |