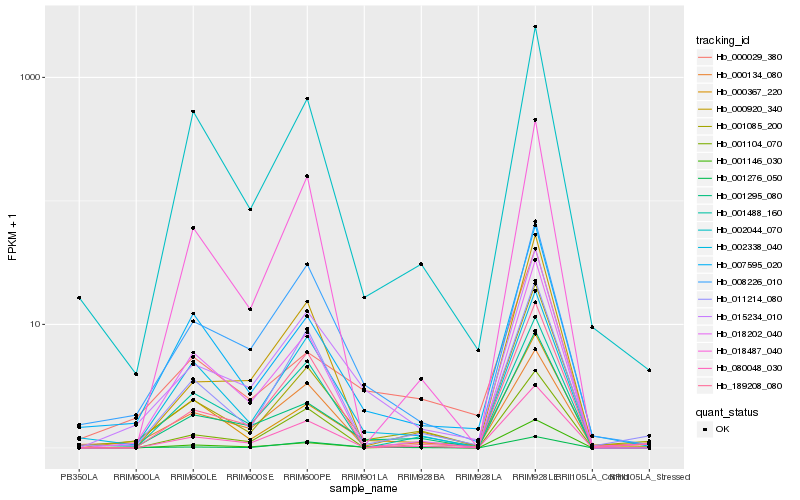
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015234_010 |
0.0 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 4B [Jatropha curcas] |
| 2 |
Hb_008226_010 |
0.1078116111 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 3 |
Hb_001276_050 |
0.127541436 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance-like protein 1O [Medicago truncatula] |
| 4 |
Hb_001295_080 |
0.1281535867 |
- |
- |
Protein KES1, putative [Ricinus communis] |
| 5 |
Hb_080048_030 |
0.1378496805 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 6 |
Hb_001085_200 |
0.1486958384 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 15 [Jatropha curcas] |
| 7 |
Hb_189208_080 |
0.1510243078 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 11-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001146_030 |
0.1517955441 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_011214_080 |
0.1531090033 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 10 |
Hb_000920_340 |
0.1561121502 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_001104_070 |
0.1566281849 |
- |
- |
hypothetical protein SORBIDRAFT_10g024640 [Sorghum bicolor] |
| 12 |
Hb_002338_040 |
0.1576200317 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007595_020 |
0.1580063072 |
- |
- |
maturase K [Hevea brasiliensis] |
| 14 |
Hb_000134_080 |
0.1602752354 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 15 |
Hb_018202_040 |
0.1625908489 |
- |
- |
PREDICTED: phosphoenolpyruvate/phosphate translocator 2, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_001488_160 |
0.1634475051 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000029_380 |
0.1637061585 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002044_070 |
0.1638203802 |
- |
- |
Chlorophyll a-b binding protein 3, chloroplastic [Theobroma cacao] |
| 19 |
Hb_018487_040 |
0.1644260309 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 20 |
Hb_000367_220 |
0.1647826192 |
- |
- |
hypothetical protein RCOM_1470580 [Ricinus communis] |