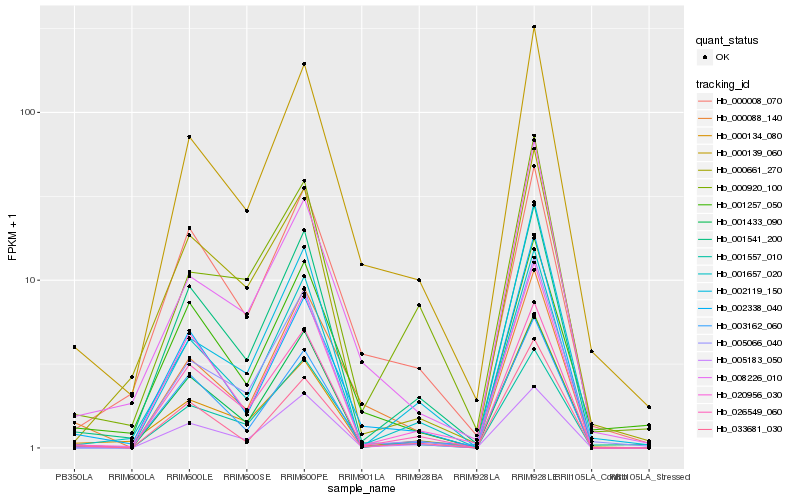
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_060 |
0.0 |
- |
- |
Photosystem I reaction center subunit II, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_008226_010 |
0.0903065354 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 3 |
Hb_001541_200 |
0.0962426293 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 4 |
Hb_001557_010 |
0.1062633572 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 5 |
Hb_005066_040 |
0.1063686565 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 6 |
Hb_000008_070 |
0.1071347928 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_001657_020 |
0.1078679357 |
- |
- |
heat shock family protein [Populus trichocarpa] |
| 8 |
Hb_001257_050 |
0.1087082265 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: protein YLS9 [Jatropha curcas] |
| 9 |
Hb_000088_140 |
0.1096437389 |
- |
- |
- |
| 10 |
Hb_000134_080 |
0.1100340798 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 11 |
Hb_002338_040 |
0.1161126548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_026549_060 |
0.1168083925 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 13 |
Hb_020956_030 |
0.1173475794 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 14 |
Hb_000920_100 |
0.1229477236 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_005183_050 |
0.1232842888 |
- |
- |
hypothetical protein RCOM_1590900 [Ricinus communis] |
| 16 |
Hb_001433_090 |
0.1302219554 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 17 |
Hb_002119_150 |
0.130403993 |
- |
- |
hypothetical protein POPTR_0008s13870g [Populus trichocarpa] |
| 18 |
Hb_003162_060 |
0.1307983535 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 19 |
Hb_000661_270 |
0.1311883765 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_033681_030 |
0.1317767266 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |