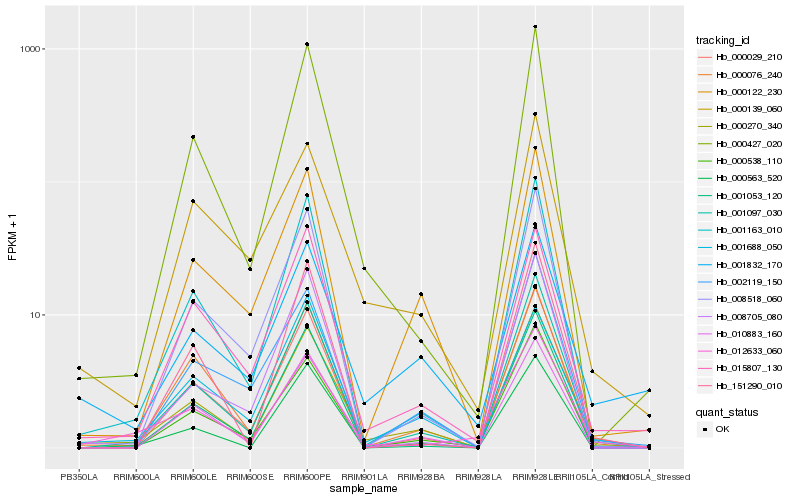
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_340 |
0.0 |
- |
- |
hypothetical protein JCGZ_11546 [Jatropha curcas] |
| 2 |
Hb_001097_030 |
0.0941735943 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 3 |
Hb_001163_010 |
0.1128164737 |
- |
- |
PREDICTED: syncoilin [Jatropha curcas] |
| 4 |
Hb_000427_020 |
0.1169234146 |
- |
- |
PsbA, partial [Dianthus pygmaeus] |
| 5 |
Hb_000538_110 |
0.1174951273 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 6 |
Hb_000076_240 |
0.1207248739 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 7 |
Hb_000122_230 |
0.1256570752 |
- |
- |
laccase, putative [Ricinus communis] |
| 8 |
Hb_000029_210 |
0.1261607285 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_010883_160 |
0.126227327 |
- |
- |
PREDICTED: pathogenesis-related protein 5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000563_520 |
0.1318016675 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 11 |
Hb_001053_120 |
0.1350019883 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46-like [Populus euphratica] |
| 12 |
Hb_012633_060 |
0.1363243747 |
- |
- |
laccase, putative [Ricinus communis] |
| 13 |
Hb_008518_060 |
0.137103387 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 1 [Jatropha curcas] |
| 14 |
Hb_002119_150 |
0.1396478353 |
- |
- |
hypothetical protein POPTR_0008s13870g [Populus trichocarpa] |
| 15 |
Hb_008705_080 |
0.1406711987 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 16 |
Hb_000139_060 |
0.1407340655 |
- |
- |
Photosystem I reaction center subunit II, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_001688_050 |
0.1413333769 |
- |
- |
PREDICTED: reticulon-like protein B9 [Jatropha curcas] |
| 18 |
Hb_001832_170 |
0.1426283314 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 19 |
Hb_015807_130 |
0.1427295525 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_151290_010 |
0.1430490149 |
- |
- |
unnamed protein product [Vitis vinifera] |