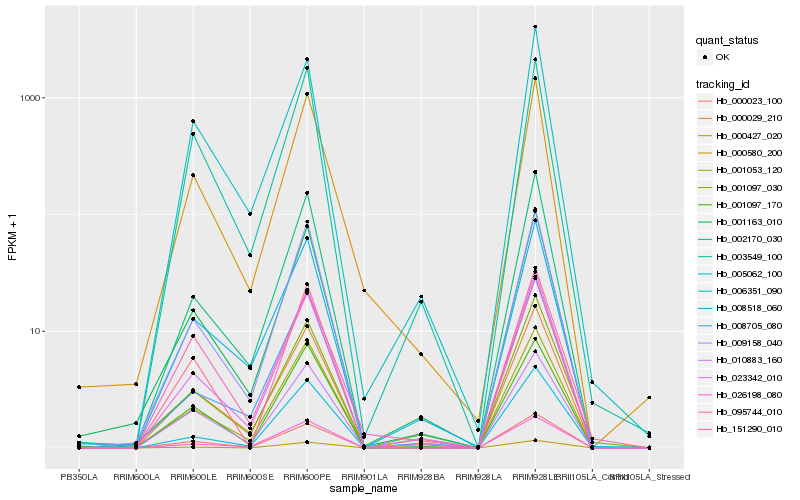
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000427_020 |
0.0 |
- |
- |
PsbA, partial [Dianthus pygmaeus] |
| 2 |
Hb_001163_010 |
0.0684772143 |
- |
- |
PREDICTED: syncoilin [Jatropha curcas] |
| 3 |
Hb_001053_120 |
0.0729648019 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46-like [Populus euphratica] |
| 4 |
Hb_009158_040 |
0.0807858194 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 5 |
Hb_023342_010 |
0.0813627857 |
- |
- |
- |
| 6 |
Hb_000029_210 |
0.0838013917 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_000023_100 |
0.0844150283 |
- |
- |
Mechanosensitive channel of small conductance-like 10 [Theobroma cacao] |
| 8 |
Hb_001097_170 |
0.0880416869 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |
| 9 |
Hb_003549_100 |
0.089959977 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |
| 10 |
Hb_002170_030 |
0.0900491083 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 11 |
Hb_008518_060 |
0.0907047563 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 1 [Jatropha curcas] |
| 12 |
Hb_095744_010 |
0.0967989852 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Fragaria vesca subsp. vesca] |
| 13 |
Hb_005062_100 |
0.1004264335 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_008705_080 |
0.1022629926 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 15 |
Hb_151290_010 |
0.1025415893 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_026198_080 |
0.102885493 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 [Populus euphratica] |
| 17 |
Hb_001097_030 |
0.1050459278 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 18 |
Hb_000580_200 |
0.1059717115 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_010883_160 |
0.1073196739 |
- |
- |
PREDICTED: pathogenesis-related protein 5 isoform X1 [Jatropha curcas] |
| 20 |
Hb_006351_090 |
0.1080081456 |
- |
- |
UDP-glycosyltransferase 85A1 [Gossypium arboreum] |